| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,101,145 – 1,101,247 |
| Length | 102 |
| Max. P | 0.684954 |

| Location | 1,101,145 – 1,101,247 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 86.77 |
| Mean single sequence MFE | -16.72 |
| Consensus MFE | -12.40 |
| Energy contribution | -13.02 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
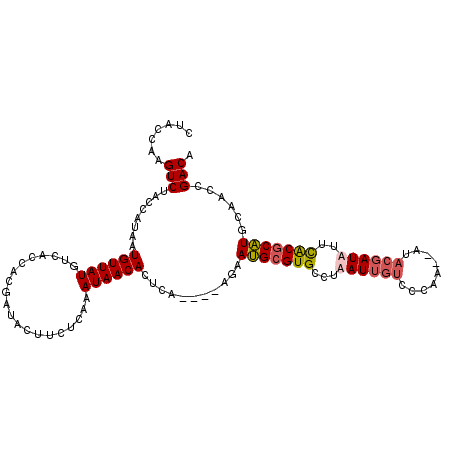
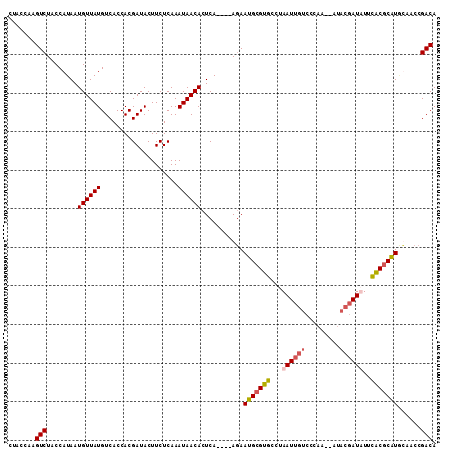
>X_DroMel_CAF1 1101145 102 - 22224390 CUACCAAGUCUACCAUAAUGUUAUGUCACCACGAUACUUCUCAAAUAACACUCACAACACAAUGAGUGCCUAAU---------UGUACGAUAUUCACGCAUGCAACCGACA .......(((.......((((.(((((.....((....))........((((((........))))))......---------.....)))))....))))......))). ( -14.32) >DroSec_CAF1 7108 107 - 1 CUACCAAGUCUACCAUAAUGUUAUGUCACCACGAUACUUCUCAAAUAACACUCA----AGAAUGCGUGCCUAAUUGUCCCAAUUGUACGAUAUUCACGCAUACAACCGACA .......(((........((((((....................))))))....----.(.(((((((....(((((.(.....).)))))...))))))).)....))). ( -16.25) >DroEre_CAF1 7137 105 - 1 CUACCAAGUCUACCAUAAUGUUAUGUCACCACGAUACUUCUCAAAUAACACUCA----AGAAUGCGUGCCUAAUUGUCCCAA--AUACGAUUUUUGCGCAUCUAACCGACA .......(((........((((((....................))))))....----(((.((((((...((((((.....--..))))))..)))))))))....))). ( -15.85) >DroYak_CAF1 7169 105 - 1 CUACCAAGUCUACCAUAAUGUUAUGUCACCACGAUACUUCUCAAAUAACACUCA----AGAACGCGUGCCUAAUUGUCCCAA--AUACGAUUUCUGCGCGUGUCACCGACA .......(((........((((((....................))))))....----.((.(((((((..((((((.....--..))))))...)))))))))...))). ( -20.45) >consensus CUACCAAGUCUACCAUAAUGUUAUGUCACCACGAUACUUCUCAAAUAACACUCA____AGAAUGCGUGCCUAAUUGUCCCAA__AUACGAUAUUCACGCAUGCAACCGACA .......(((........((((((....................))))))...........(((((((...((((((.........))))))..)))))))......))). (-12.40 = -13.02 + 0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:33 2006