
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,637,132 – 9,637,308 |
| Length | 176 |
| Max. P | 0.913496 |

| Location | 9,637,132 – 9,637,230 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -16.23 |
| Energy contribution | -17.57 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9637132 98 + 22224390 UACUGGGUAUUACUGAGCUUCACUUGGAA-CCCAAAAAAAAGAUCAAAUGCAAUUUUUUGCUACCCAGCUUCAUAACGCUUCCAAUAUGUGGGGCUGGG ...(((((....(..((.....))..).)-))))...............((((....))))..((((((((((((............)))))))))))) ( -28.40) >DroSec_CAF1 12653 95 + 1 UACAGGGUAUUUGUGAGCUCCACUUGAGA-CCCAAAAAA---AUCAAAUGCAAUUUUUUGCUACCCAGUUUCAUAACGCUCUCAAUAUGUGGAGCUGGG ....((((....(((.....))).....)-)))......---.......((((....))))..((((((((((((............)))))))))))) ( -27.50) >DroYak_CAF1 14577 92 + 1 UACAGGGUAUUACUGGGCUCCAUUUGGCAUCCCCAA--A---AUCAAAUGCAAUUUUUCGCUACCCAGUUUCAUAACUCCCUCAAUAUGUGG--CUGUA (((((((....((((((.....(((((.....))))--)---.......((........))..)))))).)).......((.(.....).))--))))) ( -17.20) >consensus UACAGGGUAUUACUGAGCUCCACUUGGAA_CCCAAAAAA___AUCAAAUGCAAUUUUUUGCUACCCAGUUUCAUAACGCUCUCAAUAUGUGG_GCUGGG ....(((.....(((((.....)))))...)))................((((....))))..((((((((((((............)))))))))))) (-16.23 = -17.57 + 1.34)

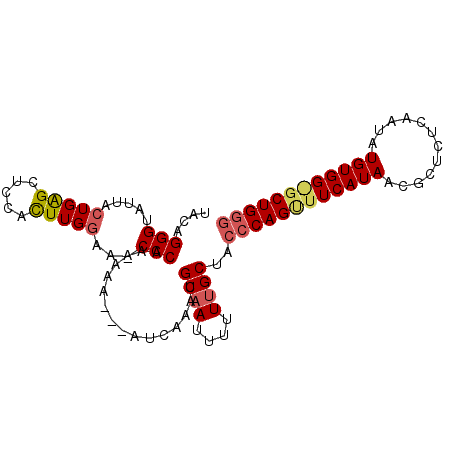
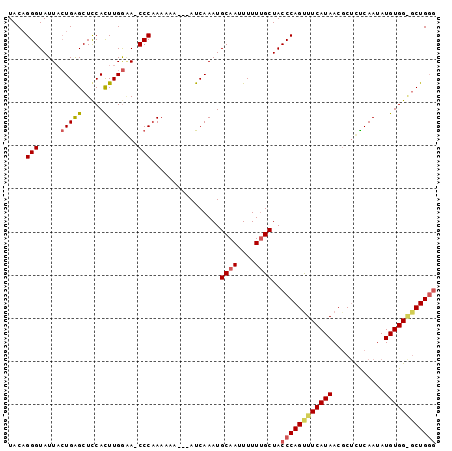
| Location | 9,637,151 – 9,637,269 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.17 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -16.46 |
| Energy contribution | -20.27 |
| Covariance contribution | 3.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9637151 118 + 22224390 UCACUUGGAA-CCCAAAAAAAAGAUCAAAUGCAAUUUUUUGCUACCCAGCUUCAUAACGCUUCCAAUAUGUGGGGCUGGGU-GCUCCGAAAGUCGCCGCAUGGCAGACCAUGUACAGACA ....(((((.-...................((((....))))((((((((((((((............)))))))))))))-).)))))..(((...((((((....))))))...))). ( -38.90) >DroSec_CAF1 12672 115 + 1 CCACUUGAGA-CCCAAAAAA---AUCAAAUGCAAUUUUUUGCUACCCAGUUUCAUAACGCUCUCAAUAUGUGGAGCUGGGU-GCUCCGAAAGUCGCCACAUGGCAGACCAUGUACAGACA ......(((.-.........---.......((((....))))((((((((((((((............)))))))))))))-)))).....(((...((((((....))))))...))). ( -34.40) >DroEre_CAF1 14129 92 + 1 CCAUGUGAGA-CCCAAA--A---AUCAAAAGCGAUUUUUUGCUACCCACUUUCAUAACUCGCUCAAUAUGUGG----------------------CCGCCUGGCAGACCAUGUACAGACA ....(((((.-......--.---......(((((....)))))..............)))))((..(((((((----------------------(.((...)).).)))))))..)).. ( -15.79) >DroYak_CAF1 14596 113 + 1 CCAUUUGGCAUCCCCAA--A---AUCAAAUGCAAUUUUUCGCUACCCAGUUUCAUAACUCCCUCAAUAUGUGG--CUGUAUGGCUCCAAAAGUCGCCGCAUGGCAGACCAUGUACAGACA ...((((((((......--.---.....))))........((((..((((..((((..........))))..)--)))..))))..)))).(((...((((((....))))))...))). ( -27.10) >consensus CCACUUGAGA_CCCAAA__A___AUCAAAUGCAAUUUUUUGCUACCCAGUUUCAUAACGCUCUCAAUAUGUGG__CUGGGU_GCUCCGAAAGUCGCCGCAUGGCAGACCAUGUACAGACA ..............................((((....)))).(((((((((((((............)))))))))))))..........(((...((((((....))))))...))). (-16.46 = -20.27 + 3.81)

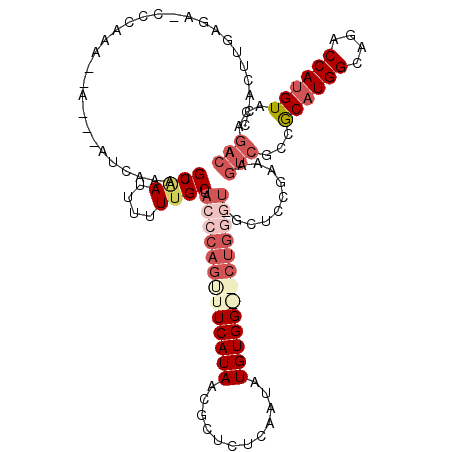

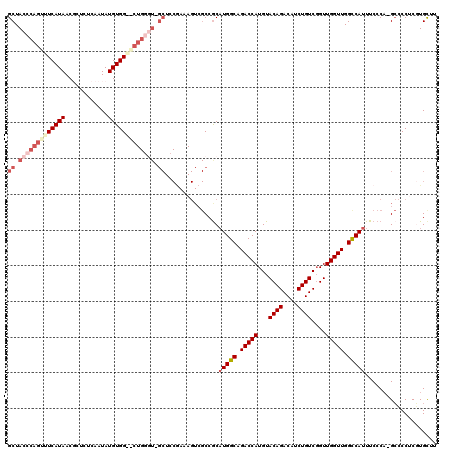
| Location | 9,637,190 – 9,637,308 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -24.44 |
| Energy contribution | -27.88 |
| Covariance contribution | 3.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9637190 118 + 22224390 GCUACCCAGCUUCAUAACGCUUCCAAUAUGUGGGGCUGGGU-GCUCCGAAAGUCGCCGCAUGGCAGACCAUGUACAGACAUCUGUCGGUUGGUUGGCCAUUUCCCA-GCCCCUCGUGUUU (((....)))..............((((((.(((((((((.-((..(....)..))...(((((.(((((...((((....))))....))))).)))))..))))-))))).)))))). ( -48.60) >DroSec_CAF1 12708 118 + 1 GCUACCCAGUUUCAUAACGCUCUCAAUAUGUGGAGCUGGGU-GCUCCGAAAGUCGCCACAUGGCAGACCAUGUACAGACAUCUGUCGGUUGGUUGGCCAUUUCCCU-GCCCCUCGUGCUU ((.(((((((((((((............)))))))))))))-((..(....)..))...(((((.(((((...((((....))))....))))).)))))......-.........)).. ( -38.90) >DroEre_CAF1 14163 97 + 1 GCUACCCACUUUCAUAACUCGCUCAAUAUGUGG----------------------CCGCCUGGCAGACCAUGUACAGACAUCUGUCCGUUGGUUGGCCAUUUCCCA-GAGCCGCUUGUUU ....................((((.....((((----------------------(((.(..((.(((.((((....))))..))).))..).)))))))......-))))......... ( -27.30) >DroYak_CAF1 14631 118 + 1 GCUACCCAGUUUCAUAACUCCCUCAAUAUGUGG--CUGUAUGGCUCCAAAAGUCGCCGCAUGGCAGACCAUGUACAGACAUCUGUCGGUUGGUUGGUCAUUUCCCAAGCCCCACUUGCUU ((((..((((..((((..........))))..)--)))..)))).......(((...((((((....))))))...)))....((.(((.((((((.......))))..)).))).)).. ( -30.10) >consensus GCUACCCAGUUUCAUAACGCUCUCAAUAUGUGG__CUGGGU_GCUCCGAAAGUCGCCGCAUGGCAGACCAUGUACAGACAUCUGUCGGUUGGUUGGCCAUUUCCCA_GCCCCUCGUGCUU ((.(((((((((((((............))))))))))))).))...............(((((.(((((...((((....))))....))))).))))).................... (-24.44 = -27.88 + 3.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:22 2006