
| Sequence ID | X_DroMel_CAF1 |
|---|---|
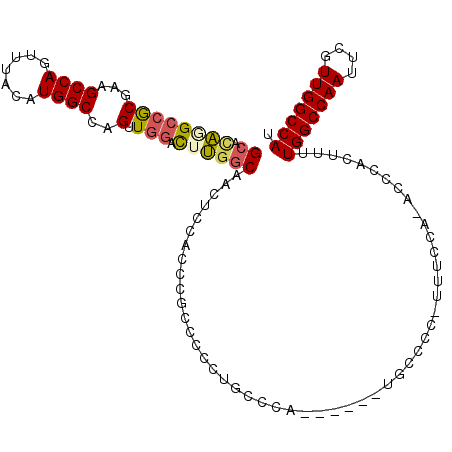
| Location | 9,619,293 – 9,619,437 |
| Length | 144 |
| Max. P | 0.884973 |

| Location | 9,619,293 – 9,619,399 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9619293 106 + 22224390 GCAUAGGCCGCUUAGCCAGUUUACAUGGCCAGUUGGACUUGGCAACUCCAAUCGCCCCCUGCCUG------UGCCCC-UUUCCAAACCCACUUUUGGCCAAUUCGUUGGCCAU ((((((((.((...((((.......))))..((((((...(....))))))).)).....)))))------)))...-................(((((((....))))))). ( -34.80) >DroSec_CAF1 79726 105 + 1 GCACAGGCCGCGAAGCCAGUUUACAUGGCCAGUUGGACUUGGCAACUCCACCCGCCCCCUUCCCA------UGCCCC-UUUCCA-ACCCACUUUUGGCCAAUUCGUUGGCCAU .....(((((((((....(....).((((((((((((...((((.....................------))))..-..))))-)).......)))))).)))).))))).. ( -30.91) >DroSim_CAF1 65332 105 + 1 GCACAGGCCGCGAAGCCAGUUUACAUGGCCAGUUGGACUUGGCAACUCCACCCGCCCCCUUCCCA------UGCCCC-UUUCCA-ACCCACUUUUGGCCAAUUCGUUGGCCAU .....(((((((((....(....).((((((((((((...((((.....................------))))..-..))))-)).......)))))).)))).))))).. ( -30.91) >DroEre_CAF1 84288 111 + 1 GCACAAGCCGCGAAGCCAGUUUACAUGGCCAGUUGGACUUGGCAACUCCACCCGCCCCCUGCCCCUGCUCCUGCCCC-UUUCCA-ACCCACUUUUGGCCAAUUCGUUGGCCAU ......((((((((....(....).((((((((((((...((((....((...((.....))...))....))))..-..))))-)).......)))))).)))).))))... ( -28.71) >DroYak_CAF1 83864 99 + 1 GCACAUGCCGCGAAGCCAGUUUACAUGGCCAGUUGGACUUGGCAACUCCACCCGCCCCCUG------------CCCC-UUUGCA-ACCCACUUUUGGCCAAUUCGUUGGCCAU ((....)).((((((...((......(((..((.(((...(....))))))..)))....)------------)..)-))))).-.........(((((((....))))))). ( -25.70) >DroAna_CAF1 101751 103 + 1 GCUCUGG--ACGAGGCCAGUUUACAUGGCCAGUUGGAU-GACCAGCACCCCUCUACCCCCUCAGU------GGACUGUUUUUUU-UUUUUUUUUUGGCCAAUUCGUUGGCCAU .....((--....(((((.......))))).(((((..-..)))))...))(((((.......))------)))..........-.........(((((((....))))))). ( -29.70) >consensus GCACAGGCCGCGAAGCCAGUUUACAUGGCCAGUUGGACUUGGCAACUCCACCCGCCCCCUGCCCA______UGCCCC_UUUCCA_ACCCACUUUUGGCCAAUUCGUUGGCCAU ((.((((((((...((((.......))))..).))).))))))...................................................(((((((....))))))). (-21.06 = -21.32 + 0.25)

| Location | 9,619,293 – 9,619,399 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -38.32 |
| Consensus MFE | -24.33 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9619293 106 - 22224390 AUGGCCAACGAAUUGGCCAAAAGUGGGUUUGGAAA-GGGGCA------CAGGCAGGGGGCGAUUGGAGUUGCCAAGUCCAACUGGCCAUGUAAACUGGCUAAGCGGCCUAUGC .(((((((....)))))))..(((.((((((.(..-..(((.------(((...(((((((((....))))))...)))..)))))).).)))))).)))..(((.....))) ( -36.70) >DroSec_CAF1 79726 105 - 1 AUGGCCAACGAAUUGGCCAAAAGUGGGU-UGGAAA-GGGGCA------UGGGAAGGGGGCGGGUGGAGUUGCCAAGUCCAACUGGCCAUGUAAACUGGCUUCGCGGCCUGUGC .(((((((....)))))))...(..(((-(((((.-((.(((------(((..((.((((...(((.....))).))))..))..))))))...))...))).)))))..).. ( -40.20) >DroSim_CAF1 65332 105 - 1 AUGGCCAACGAAUUGGCCAAAAGUGGGU-UGGAAA-GGGGCA------UGGGAAGGGGGCGGGUGGAGUUGCCAAGUCCAACUGGCCAUGUAAACUGGCUUCGCGGCCUGUGC .(((((((....)))))))...(..(((-(((((.-((.(((------(((..((.((((...(((.....))).))))..))..))))))...))...))).)))))..).. ( -40.20) >DroEre_CAF1 84288 111 - 1 AUGGCCAACGAAUUGGCCAAAAGUGGGU-UGGAAA-GGGGCAGGAGCAGGGGCAGGGGGCGGGUGGAGUUGCCAAGUCCAACUGGCCAUGUAAACUGGCUUCGCGGCUUGUGC .(((((((....)))))))...(..(((-((((..-.((.(((..(((.((.(((.((((...(((.....))).))))..))).)).)))...))).)))).)))))..).. ( -42.50) >DroYak_CAF1 83864 99 - 1 AUGGCCAACGAAUUGGCCAAAAGUGGGU-UGCAAA-GGGG------------CAGGGGGCGGGUGGAGUUGCCAAGUCCAACUGGCCAUGUAAACUGGCUUCGCGGCAUGUGC .(((((((....))))))).((((.(((-((((..-.((.------------(((.((((...(((.....))).))))..))).)).))).)))).))))............ ( -35.80) >DroAna_CAF1 101751 103 - 1 AUGGCCAACGAAUUGGCCAAAAAAAAAA-AAAAAAACAGUCC------ACUGAGGGGGUAGAGGGGUGCUGGUC-AUCCAACUGGCCAUGUAAACUGGCCUCGU--CCAGAGC .(((((((....))))))).........-.............------.((((.(((((...((..((((((((-(......)))))).)))..)).))))).)--.)))... ( -34.50) >consensus AUGGCCAACGAAUUGGCCAAAAGUGGGU_UGGAAA_GGGGCA______UGGGAAGGGGGCGGGUGGAGUUGCCAAGUCCAACUGGCCAUGUAAACUGGCUUCGCGGCCUGUGC .(((((((....)))))))..................((((...............((((...(((.....))).))))....(((((.(....)))))).....)))).... (-24.33 = -25.00 + 0.67)



| Location | 9,619,329 – 9,619,437 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -18.22 |
| Energy contribution | -18.13 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9619329 108 + 22224390 ACUUGGCAACUCCAAUCGCCCCCUGCCUG------UGCCCC-UUUCCAAACCCACUUUUGGCCAAUUCGUUGGCCAUUUUAAUGGCCGCCGACCGAACGUCGCCGUUUCG--UACUG ....((((...............)))).(------(((...-...(((((......))))).....(((.(((((((....))))))).)))..(((((....))))).)--))).. ( -24.36) >DroSec_CAF1 79762 107 + 1 ACUUGGCAACUCCACCCGCCCCCUUCCCA------UGCCCC-UUUCCA-ACCCACUUUUGGCCAAUUCGUUGGCCAUUUUAAUGGCCGCCGACCGAACGUCGCCGUUUCG--UACUU ....(((..........((..........------.))...-......-....((.(((((.....(((.(((((((....))))))).)))))))).)).)))......--..... ( -20.90) >DroSim_CAF1 65368 107 + 1 ACUUGGCAACUCCACCCGCCCCCUUCCCA------UGCCCC-UUUCCA-ACCCACUUUUGGCCAAUUCGUUGGCCAUUUUAAUGGCCGCCGACCGAACGUCGCCGUUUCG--UAGUU ....(((..........((..........------.))...-......-....((.(((((.....(((.(((((((....))))))).)))))))).)).)))......--..... ( -20.90) >DroEre_CAF1 84324 115 + 1 ACUUGGCAACUCCACCCGCCCCCUGCCCCUGCUCCUGCCCC-UUUCCA-ACCCACUUUUGGCCAAUUCGUUGGCCAUUUUAAUGGCCGCCGACCGAACGUCGCCGUUCCUUCUUCUU ..(((((..........((.....))....((....))...-......-...........))))).(((.(((((((....))))))).)))..(((((....)))))......... ( -24.90) >DroYak_CAF1 83900 103 + 1 ACUUGGCAACUCCACCCGCCCCCUG------------CCCC-UUUGCA-ACCCACUUUUGGCCAAUUCGUUGGCCAUUUUAAUGGCCGCCGACCGAACGUCGCCGUUUCUUCUACUU ....(((................((------------(...-...)))-....((.(((((.....(((.(((((((....))))))).)))))))).)).)))............. ( -23.50) >DroAna_CAF1 101785 105 + 1 AU-GACCAGCACCCCUCUACCCCCUCAGU------GGACUGUUUUUUU-UUUUUUUUUUGGCCAAUUCGUUGGCCAUUUUAAUGGCCGGCGACCAAACGUCGACGUU--G--UGCUC ..-....(((((...(((((.......))------)))..........-.........(((.....(((((((((((....))))))))))))))((((....))))--)--)))). ( -29.20) >consensus ACUUGGCAACUCCACCCGCCCCCUGCCCA______UGCCCC_UUUCCA_ACCCACUUUUGGCCAAUUCGUUGGCCAUUUUAAUGGCCGCCGACCGAACGUCGCCGUUUCG__UACUU ....(((.((.........................................((......)).....(((..((((((....))))))..)))......)).)))............. (-18.22 = -18.13 + -0.08)


| Location | 9,619,329 – 9,619,437 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -35.13 |
| Consensus MFE | -25.33 |
| Energy contribution | -25.55 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9619329 108 - 22224390 CAGUA--CGAAACGGCGACGUUCGGUCGGCGGCCAUUAAAAUGGCCAACGAAUUGGCCAAAAGUGGGUUUGGAAA-GGGGCA------CAGGCAGGGGGCGAUUGGAGUUGCCAAGU .....--......((((((.((((((((.(.(((.......(((((((....)))))))...(((..(((....)-))..))------).)))....).)))))))))))))).... ( -40.40) >DroSec_CAF1 79762 107 - 1 AAGUA--CGAAACGGCGACGUUCGGUCGGCGGCCAUUAAAAUGGCCAACGAAUUGGCCAAAAGUGGGU-UGGAAA-GGGGCA------UGGGAAGGGGGCGGGUGGAGUUGCCAAGU .....--......((((((.((((.(((.(((((((....))))))......(..(((.......)))-..)...-......------.........).))).)))))))))).... ( -34.30) >DroSim_CAF1 65368 107 - 1 AACUA--CGAAACGGCGACGUUCGGUCGGCGGCCAUUAAAAUGGCCAACGAAUUGGCCAAAAGUGGGU-UGGAAA-GGGGCA------UGGGAAGGGGGCGGGUGGAGUUGCCAAGU .....--......((((((.((((.(((.(((((((....))))))......(..(((.......)))-..)...-......------.........).))).)))))))))).... ( -34.30) >DroEre_CAF1 84324 115 - 1 AAGAAGAAGGAACGGCGACGUUCGGUCGGCGGCCAUUAAAAUGGCCAACGAAUUGGCCAAAAGUGGGU-UGGAAA-GGGGCAGGAGCAGGGGCAGGGGGCGGGUGGAGUUGCCAAGU .....((..(((((....)))))..))(((((((((....))))))(((..(((.(((........((-(.....-..)))....((....))....))).)))...)))))).... ( -35.40) >DroYak_CAF1 83900 103 - 1 AAGUAGAAGAAACGGCGACGUUCGGUCGGCGGCCAUUAAAAUGGCCAACGAAUUGGCCAAAAGUGGGU-UGCAAA-GGGG------------CAGGGGGCGGGUGGAGUUGCCAAGU .............((((((.((((.(((.(..(((((....(((((((....)))))))..))))).(-(((...-...)------------)))..).))).)))))))))).... ( -36.30) >DroAna_CAF1 101785 105 - 1 GAGCA--C--AACGUCGACGUUUGGUCGCCGGCCAUUAAAAUGGCCAACGAAUUGGCCAAAAAAAAAA-AAAAAAACAGUCC------ACUGAGGGGGUAGAGGGGUGCUGGUC-AU .((((--(--..((.((((.....)))).))(((.......(((((((....))))))).........-.......(((...------.)))....)))......)))))....-.. ( -30.10) >consensus AAGUA__CGAAACGGCGACGUUCGGUCGGCGGCCAUUAAAAUGGCCAACGAAUUGGCCAAAAGUGGGU_UGGAAA_GGGGCA______UGGGAAGGGGGCGGGUGGAGUUGCCAAGU .............((((((.((((.(((.(((((((....)))))).........(((....................)))................).))).)))))))))).... (-25.33 = -25.55 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:13 2006