
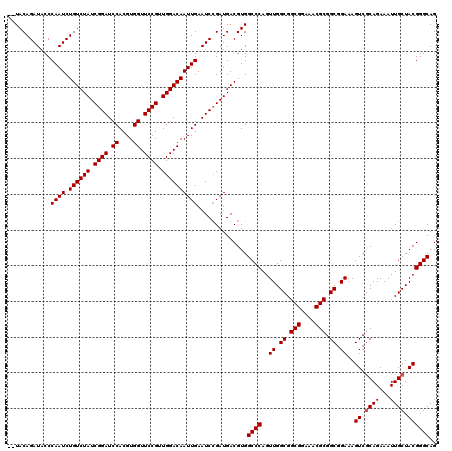
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,618,108 – 9,618,287 |
| Length | 179 |
| Max. P | 0.973362 |

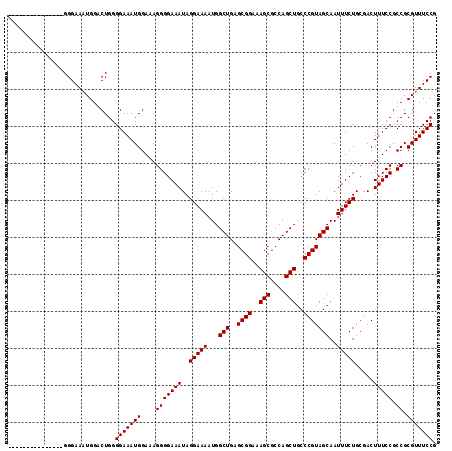
| Location | 9,618,108 – 9,618,226 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.63 |
| Mean single sequence MFE | -43.15 |
| Consensus MFE | -41.92 |
| Energy contribution | -42.18 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9618108 118 + 22224390 --UACAGAUACCCAAUCUGUCUAUCGGAUCCACGUGGUUCCGUUGGACAAUUGAAUCAGAUGACGUGGCCCAGUUGGCGGCGGAAACGCGGCGGAAAGUCGCAGAAAUUGCUACGGGCAG --....(((...((((.((((((.((((.((....)).)))).)))))))))).)))..........((((..((.((.(((....))).)).))..((.((((...)))).)))))).. ( -43.80) >DroSec_CAF1 77451 118 + 1 --UACAGAUACCCAAUCUGUCUAUCGGAUCCACGUGGUUCCGUUGGACAAUUGAAUCCGAUGACGUGGCCCAGUUGGCGGCGGAAACGCGGCGGAAAGUCUCAGAAAUUGCUACGGGCAG --(((((((.....))))(((.(((((((.((....((((....))))...)).)))))))))))))((((.((.(((((((....)))(((.....)))........)))))))))).. ( -40.00) >DroEre_CAF1 83024 120 + 1 GAUACAGAUACCCAAUCUGUCUAUCGGAUCCACGUGGUUCCGUUGGACAAUUGAAUCCGAUGACGUGGCCCAGUUGGCGGCGGAAACGCGGCGGAAAGUCGCAGAAAUUGCUACGGGCAG ..(((((((.....))))(((.(((((((.((....((((....))))...)).)))))))))))))((((..((.((.(((....))).)).))..((.((((...)))).)))))).. ( -43.60) >DroYak_CAF1 82499 120 + 1 GAUACAGAUACCCAAUCUGUCUAUCGGAUCCACGUGGUUCCGCUGGACAAUUGAAUCCGAUGACGUGGCCCAGUUGGCGGCGGAAACGCGGCGGAAAGUCGCAGAAAUUGCUACGGGCAG ..(((((((.....))))(((.((((((((((.(((....)))))))........))))))))))))((((..((.((.(((....))).)).))..((.((((...)))).)))))).. ( -45.20) >consensus __UACAGAUACCCAAUCUGUCUAUCGGAUCCACGUGGUUCCGUUGGACAAUUGAAUCCGAUGACGUGGCCCAGUUGGCGGCGGAAACGCGGCGGAAAGUCGCAGAAAUUGCUACGGGCAG ............((((.((((((.((((.((....)).)))).))))))))))..............((((..((.((.(((....))).)).))..((.((((...)))).)))))).. (-41.92 = -42.18 + 0.25)


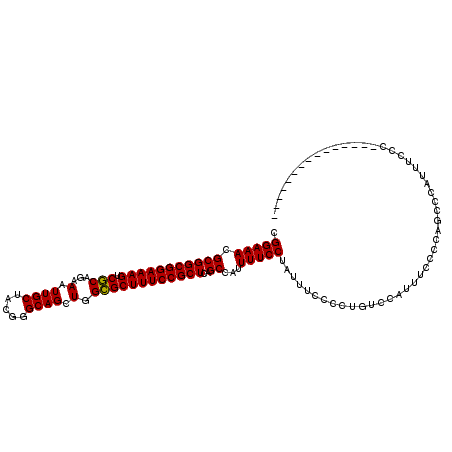
| Location | 9,618,186 – 9,618,287 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 90.02 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -27.83 |
| Energy contribution | -27.45 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9618186 101 + 22224390 CGGAAACGCGGCGGAAAGUCGCAGAAAUUGCUACGGGCAGCUGGCGCUUUCCGCUCAGCCAUUUUCCUAUUUCCCCUUUCCAUUUCCCCAGUCCAUUUCCC--------------- .(((((.(((((((((((.(((...(.((((.....)))).).))))))))))))..))...)))))..................................--------------- ( -28.00) >DroSec_CAF1 77529 101 + 1 CGGAAACGCGGCGGAAAGUCUCAGAAAUUGCUACGGGCAGCUGGGGCUUUCCGCUCAGCCAUUUUCCUAUUUCCCCUUUCCAUUUCCCCAGUCCAUUUCCC--------------- .(((((.(((((((((((((((((....(((.....))).)))))))))))))))..))...)))))..................................--------------- ( -33.80) >DroEre_CAF1 83104 101 + 1 CGGAAACGCGGCGGAAAGUCGCAGAAAUUGCUACGGGCAGCUGGCGCUUUCCGCUCAGCCAUUUUCCUAUUUCCCCUGUCCAUUUCCCCAGCCCAUUUCCC--------------- .(((((.(.((((((((...((((....(((.....)))(((((((.....))).))))................))))...)))))...)))).))))).--------------- ( -30.40) >DroYak_CAF1 82579 116 + 1 CGGAAACGCGGCGGAAAGUCGCAGAAAUUGCUACGGGCAGCUGGCGCUUUCCGCUCAGCCAUUUUCCUAUUUCCCCUGUCCAUUUCCCCAGCCCACUGCCCAUUUCCCAUUUUUUC .(((((.(.(((((...((.((((...)))).))((((.(((((((.....))).)))).................((..........)))))).)))))).)))))......... ( -32.40) >consensus CGGAAACGCGGCGGAAAGUCGCAGAAAUUGCUACGGGCAGCUGGCGCUUUCCGCUCAGCCAUUUUCCUAUUUCCCCUGUCCAUUUCCCCAGCCCAUUUCCC_______________ .(((((.(((((((((((.(((...(.((((.....)))).).))))))))))))..))...)))))................................................. (-27.83 = -27.45 + -0.38)


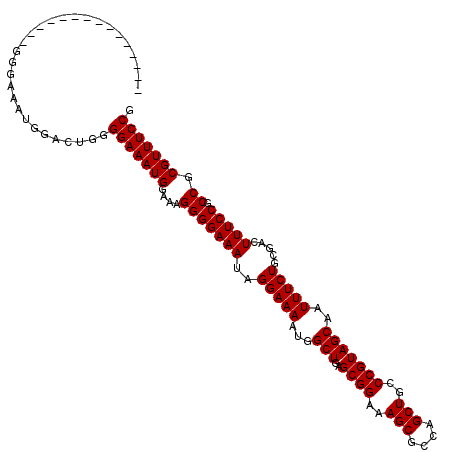
| Location | 9,618,186 – 9,618,287 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 90.02 |
| Mean single sequence MFE | -35.16 |
| Consensus MFE | -31.40 |
| Energy contribution | -31.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9618186 101 - 22224390 ---------------GGGAAAUGGACUGGGGAAAUGGAAAGGGGAAAUAGGAAAAUGGCUGAGCGGAAAGCGCCAGCUGCCCGUAGCAAUUUCUGCGACUUUCCGCCGCGUUUCCG ---------------.(((((((...((((((((.(....(.((((((........(((((.(((.....))))))))((.....)).)))))).)..)))))).)))))))))). ( -33.50) >DroSec_CAF1 77529 101 - 1 ---------------GGGAAAUGGACUGGGGAAAUGGAAAGGGGAAAUAGGAAAAUGGCUGAGCGGAAAGCCCCAGCUGCCCGUAGCAAUUUCUGAGACUUUCCGCCGCGUUUCCG ---------------.(((((((...(((.....(((((((........(((((...(((..((((..(((....)))..)))))))..)))))....))))))))))))))))). ( -30.70) >DroEre_CAF1 83104 101 - 1 ---------------GGGAAAUGGGCUGGGGAAAUGGACAGGGGAAAUAGGAAAAUGGCUGAGCGGAAAGCGCCAGCUGCCCGUAGCAAUUUCUGCGACUUUCCGCCGCGUUUCCG ---------------.((((((((((..(((((.(.(.(((..(............(((((.(((.....))))))))((.....))..)..)))).).)))))))).))))))). ( -36.70) >DroYak_CAF1 82579 116 - 1 GAAAAAAUGGGAAAUGGGCAGUGGGCUGGGGAAAUGGACAGGGGAAAUAGGAAAAUGGCUGAGCGGAAAGCGCCAGCUGCCCGUAGCAAUUUCUGCGACUUUCCGCCGCGUUUCCG .........((((((((((.(..((.((.((((((.(..(.(((............(((((.(((.....)))))))).))).)..).)))))).)).))..).))).))))))). ( -39.72) >consensus _______________GGGAAAUGGACUGGGGAAAUGGAAAGGGGAAAUAGGAAAAUGGCUGAGCGGAAAGCGCCAGCUGCCCGUAGCAAUUUCUGCGACUUUCCGCCGCGUUUCCG .............................(((((((....(((((((..(((((...(((..((((..(((....)))..)))))))..))))).....))))).)).))))))). (-31.40 = -31.40 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:10 2006