
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,614,684 – 9,614,858 |
| Length | 174 |
| Max. P | 1.000000 |

| Location | 9,614,684 – 9,614,786 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 70.00 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -16.78 |
| Energy contribution | -16.46 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.74 |
| SVM decision value | 7.34 |
| SVM RNA-class probability | 1.000000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9614684 102 + 22224390 CAUUUUGCCAGCUGGCGCAGAGAAAAAACCAAAAAAAAGAAAAAUACUAAUAAAAAAAAGGGAAACUUGCAGUUGGCAAUUACUUGGCCUAAACCAAGUAAU ....((((((((((.(.....)....................................(((....))).))))))))))((((((((......)))))))). ( -26.10) >DroSim_CAF1 60919 92 + 1 CAUUUUGCCAGCUGGUGCAGAGAAAAAAUAAAAAA-AACCAAAA---------AAAAAAGGGAAACUUGCAGUUGGCAAUUACUUGGCCUAAACCAAGUAAU ....((((((((((((...................-.)))....---------.....(((....)))..)))))))))((((((((......)))))))). ( -26.45) >DroAna_CAF1 95817 86 + 1 CAUUUUUUCAGUUGGCCCUAA---AAAAUGGAGGA-A------------AGAAAAAAAAGGGAAAAAUCCGAGUGGCAAUUACUUGGUCUAAGCCGGAUAAU ..(((((((..((..((....---.....))..))-.------------.)))))))...((......(((((((.....)))))))......))....... ( -15.40) >consensus CAUUUUGCCAGCUGGCGCAGAGAAAAAAUAAAAAA_AA__AAAA_____A_AAAAAAAAGGGAAACUUGCAGUUGGCAAUUACUUGGCCUAAACCAAGUAAU ....((((((((((.......................................................))))))))))((((((((......)))))))). (-16.78 = -16.46 + -0.32)



| Location | 9,614,684 – 9,614,786 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 70.00 |
| Mean single sequence MFE | -17.33 |
| Consensus MFE | -10.72 |
| Energy contribution | -11.73 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9614684 102 - 22224390 AUUACUUGGUUUAGGCCAAGUAAUUGCCAACUGCAAGUUUCCCUUUUUUUUAUUAGUAUUUUUCUUUUUUUUGGUUUUUUCUCUGCGCCAGCUGGCAAAAUG .(((((((((....)))))))))((((((.((((.((..............(..((.......))..)..............))..).))).)))))).... ( -20.49) >DroSim_CAF1 60919 92 - 1 AUUACUUGGUUUAGGCCAAGUAAUUGCCAACUGCAAGUUUCCCUUUUUU---------UUUUGGUU-UUUUUUAUUUUUUCUCUGCACCAGCUGGCAAAAUG .(((((((((....)))))))))((((((.(((((((............---------.))))((.-.................))..))).)))))).... ( -19.59) >DroAna_CAF1 95817 86 - 1 AUUAUCCGGCUUAGACCAAGUAAUUGCCACUCGGAUUUUUCCCUUUUUUUUCU------------U-UCCUCCAUUUU---UUAGGGCCAACUGAAAAAAUG .......(((....((...))....)))....(((....)))...........------------.-.......((((---((((......))))))))... ( -11.90) >consensus AUUACUUGGUUUAGGCCAAGUAAUUGCCAACUGCAAGUUUCCCUUUUUUUU_U_____UUUU__UU_UUUUUCAUUUUUUCUCUGCGCCAGCUGGCAAAAUG .((((((((......))))))))((((((.(((.......................................................))).)))))).... (-10.72 = -11.73 + 1.00)

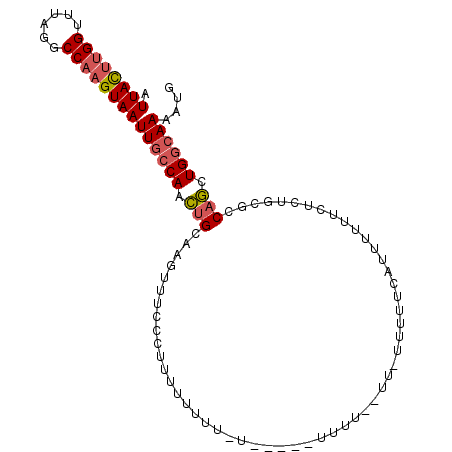

| Location | 9,614,720 – 9,614,826 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -13.39 |
| Energy contribution | -14.32 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.63 |
| SVM decision value | 4.62 |
| SVM RNA-class probability | 0.999930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9614720 106 + 22224390 AAGAAAAAUACUAAUAAAAAAAAGGGAAACUUGCAGUUGGCAAUUACUUGGCCUAAACCAAGUAAUGAUAAAUAAUUAAGGCAAAACAGCGCCAGAUUAAUAUGCA ......................(((....)))(((.(((((.(((((((((......)))))))))..............((......))))))).......))). ( -23.80) >DroSim_CAF1 60954 97 + 1 AACCAAAA---------AAAAAAGGGAAACUUGCAGUUGGCAAUUACUUGGCCUAAACCAAGUAAUGAUAAAUAAUUAAGGCAAAACAGCGCCAGAUUAAUAUGCA ........---------.....(((....)))(((.(((((.(((((((((......)))))))))..............((......))))))).......))). ( -23.80) >DroEre_CAF1 78329 88 + 1 ------------------CAAAAGGGAAACUUGCGGCUGGCAAUUACUUGGCCUAAAGCAAGUAAUGAUAAAUAAUUAAGGCAAAACAGCGCCAGAUUAAUAUGCA ------------------....(((....)))(((.(((((.((((((((........))))))))..............((......))))))).......))). ( -22.10) >DroAna_CAF1 95849 86 + 1 A------------AGAAAAAAAAGGGAAAAAUCCGAGUGGCAAUUACUUGGUCUAAGCCGGAUAAUGAUUUAUAAUUAAGCCAG--------UAGAUUAAUAUGCA .------------...........((......(((((((.....)))))))......))(.(((.((((((((..........)--------))))))).))).). ( -15.50) >consensus A________________AAAAAAGGGAAACUUGCAGUUGGCAAUUACUUGGCCUAAACCAAGUAAUGAUAAAUAAUUAAGGCAAAACAGCGCCAGAUUAAUAUGCA ......................(((....))).......((((((((((((......)))))))))......(((((..(((........))).)))))...))). (-13.39 = -14.32 + 0.94)


| Location | 9,614,746 – 9,614,858 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -23.74 |
| Energy contribution | -23.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9614746 112 + 22224390 AAACUUGCAGUUGGCAAUUACUUGGCCUAAACCAAGUAAUGAUAAAUAAUUAAGGCAAAACAGCGCCAGAUUAAUAUGCAUUUAUUCACACUUUGGCGGGGGAG-----CCAGUGAA .....((((.(((((.(((((((((......)))))))))..............((......))))))).......))))....(((((....((((......)-----)))))))) ( -30.40) >DroSec_CAF1 73584 112 + 1 AAACUUGCAGUUGGCAAUUACUUGGCCUAAACCAAGUAAUGAUAAAUAAUUAAGGCAAAACAGCGCCAGAUUAAUAUGCAUUUAUUCACACUUUGGCGGGGGAG-----CCAGUGAA .....((((.(((((.(((((((((......)))))))))..............((......))))))).......))))....(((((....((((......)-----)))))))) ( -30.40) >DroSim_CAF1 60971 112 + 1 AAACUUGCAGUUGGCAAUUACUUGGCCUAAACCAAGUAAUGAUAAAUAAUUAAGGCAAAACAGCGCCAGAUUAAUAUGCAUUUAUUCACACUUUGGCGGGGGAG-----CCAAGGAA .....((((.(((((.(((((((((......)))))))))..............((......))))))).......))))..........(((((((......)-----)))))).. ( -32.30) >DroEre_CAF1 78337 117 + 1 AAACUUGCGGCUGGCAAUUACUUGGCCUAAAGCAAGUAAUGAUAAAUAAUUAAGGCAAAACAGCGCCAGAUUAAUAUGCAUUUAUUCACACUUUGGCGGGGGAGCCGAGCCAGUGAA .........((((((.((((((((........)))))))).............(((....(..(((((((..((((......)))).....)))))))..)..)))..))))))... ( -31.90) >DroYak_CAF1 78444 115 + 1 AAACUUGCAGUUGGCAAUUACUUGGCCUAAACCAAGUAAUGAUAAAUAAUUAAGGCAAAACAGCGCCAGAUUAAUAUGCAUUUAUUCACACUUUGGCGGGGGAGC--AGCCAGCGAA .........((((((.(((((((((......)))))))))..............((....(..(((((((..((((......)))).....)))))))..)..))--.))))))... ( -31.40) >consensus AAACUUGCAGUUGGCAAUUACUUGGCCUAAACCAAGUAAUGAUAAAUAAUUAAGGCAAAACAGCGCCAGAUUAAUAUGCAUUUAUUCACACUUUGGCGGGGGAG_____CCAGUGAA ...(((((...((((.(((((((((......)))))))))..............((......))))))((.(((.......))).))........)))))((.......))...... (-23.74 = -23.94 + 0.20)



| Location | 9,614,746 – 9,614,858 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9614746 112 - 22224390 UUCACUGG-----CUCCCCCGCCAAAGUGUGAAUAAAUGCAUAUUAAUCUGGCGCUGUUUUGCCUUAAUUAUUUAUCAUUACUUGGUUUAGGCCAAGUAAUUGCCAACUGCAAGUUU ((((((((-----(......))))....)))))....((((........((((((......))..............((((((((((....)))))))))).))))..))))..... ( -28.00) >DroSec_CAF1 73584 112 - 1 UUCACUGG-----CUCCCCCGCCAAAGUGUGAAUAAAUGCAUAUUAAUCUGGCGCUGUUUUGCCUUAAUUAUUUAUCAUUACUUGGUUUAGGCCAAGUAAUUGCCAACUGCAAGUUU ((((((((-----(......))))....)))))....((((........((((((......))..............((((((((((....)))))))))).))))..))))..... ( -28.00) >DroSim_CAF1 60971 112 - 1 UUCCUUGG-----CUCCCCCGCCAAAGUGUGAAUAAAUGCAUAUUAAUCUGGCGCUGUUUUGCCUUAAUUAUUUAUCAUUACUUGGUUUAGGCCAAGUAAUUGCCAACUGCAAGUUU ....((((-----(......)))))((((((........))))))....((((((......))..............((((((((((....)))))))))).))))........... ( -27.50) >DroEre_CAF1 78337 117 - 1 UUCACUGGCUCGGCUCCCCCGCCAAAGUGUGAAUAAAUGCAUAUUAAUCUGGCGCUGUUUUGCCUUAAUUAUUUAUCAUUACUUGCUUUAGGCCAAGUAAUUGCCAGCCGCAAGUUU ....(((((..(((.....(((((.((((((........))))))....))))).......))).............(((((((((.....).)))))))).))))).(....)... ( -29.10) >DroYak_CAF1 78444 115 - 1 UUCGCUGGCU--GCUCCCCCGCCAAAGUGUGAAUAAAUGCAUAUUAAUCUGGCGCUGUUUUGCCUUAAUUAUUUAUCAUUACUUGGUUUAGGCCAAGUAAUUGCCAACUGCAAGUUU ...((((((.--((.....(((((.((((((........))))))....))))).......))..............((((((((((....)))))))))).))))...))...... ( -28.30) >consensus UUCACUGG_____CUCCCCCGCCAAAGUGUGAAUAAAUGCAUAUUAAUCUGGCGCUGUUUUGCCUUAAUUAUUUAUCAUUACUUGGUUUAGGCCAAGUAAUUGCCAACUGCAAGUUU ....................((...((((((........))))))....((((((......))..............((((((((((....)))))))))).))))...))...... (-21.70 = -21.90 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:07 2006