
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,600,436 – 9,600,563 |
| Length | 127 |
| Max. P | 0.986869 |

| Location | 9,600,436 – 9,600,536 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.29 |
| Mean single sequence MFE | -40.37 |
| Consensus MFE | -15.18 |
| Energy contribution | -15.60 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.38 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
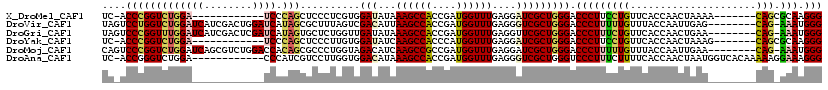
>X_DroMel_CAF1 9600436 100 + 22224390 UC-ACCCGGUCUGGA------------UCCCAGCUCCCUCGUGGAUAUAAAGCCACCGAUGGUUUGAGGAUCGCUGGGACCCUUCCUGUUCACCAACUAAAA-------CAGCGCAAGGG ..-.((((((...((------------((((((..((.((((((........))).))).)).))).))))))))))).(((((((((((..........))-------))).).))))) ( -35.50) >DroVir_CAF1 88494 111 + 1 UAGUCCUGGUCUGGAUCAUCGACUGGAUCAUAGCGCUUUAGUCGACAUUAAGCCACCGAUGGUUUGAGGGUCGCUGGGACCCUUUUUGUUUACCAAUUGAG--------CAG-AAAUGGG ..((((..((...((((.(((((((((.(.....).)))))))))..((((((((....)))))))).))))))..))))((((((((((((.....))))--------)))-))).)). ( -39.50) >DroGri_CAF1 71695 111 + 1 UAGUCCCGGUUUGGAUCAUCGACUCGAUCAUAGUGCUCUGGUUGAUAUAAAGCCACCGAUGGUUUGAGGUUCGCUGGGACCCUUUCUGUUCACCAACUGAA--------CAG-AAAUGGG ..((((((((..((((((((((((.((.(.....).)).)))))))...((((((....))))))..)))))))))))))((((((((((((.....))))--------)))-))).)). ( -43.50) >DroYak_CAF1 62699 100 + 1 UC-ACCCGGUCUGGA------------UCCCAGCUCCCUUGUGGAUAUCAAGCCACCCAUGGUUUGAGGAUCGCUGGGACCCUUCCUGUUCACCAACUAAAG-------CAGCGCAAGGG ..-.((((((...((------------(((......((....))...((((((((....))))))))))))))))))).(((((((((((..........))-------))).).))))) ( -35.40) >DroMoj_CAF1 84606 111 + 1 CAGUCCCGGUCUGGAUCAGCGUCUGGACCACAGCGCCCUGGUAGACAUCAAGCCGCCGAUGGUUUGAGGAUCGCUGGGACCCUUUUUGUUUACCAAUUGAA--------CAG-AAAUGGG ..(((((((((..(((....)))..))))..((((.((((.....))((((((((....))))))))))..)))))))))((((((((((((.....))))--------)))-))).)). ( -44.40) >DroAna_CAF1 74670 107 + 1 UC-ACCGGGUCUGGA------------CCCAUCGUCCUUGGUGGACAUAAAGCCACCGAUGGUUUGAGGGUCGCUGGGUCCCUUUCUUUUCACCAACUAAUGGUCACAAAAAGGAAAGGG ..-(((.(((...((------------(((.(((.(((((((((........))))))).))..))))))))))).)))(((((((((((.((((.....)))).....))))))))))) ( -43.90) >consensus UA_ACCCGGUCUGGA____________UCACAGCGCCCUGGUGGACAUAAAGCCACCGAUGGUUUGAGGAUCGCUGGGACCCUUUCUGUUCACCAACUAAA________CAG_AAAAGGG ....(((((((((((((........)))).)))..........(((...((((((....))))))....))))))))).(((((((((.....................))).))).))) (-15.18 = -15.60 + 0.42)
| Location | 9,600,463 – 9,600,563 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 86.31 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -24.48 |
| Energy contribution | -24.23 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
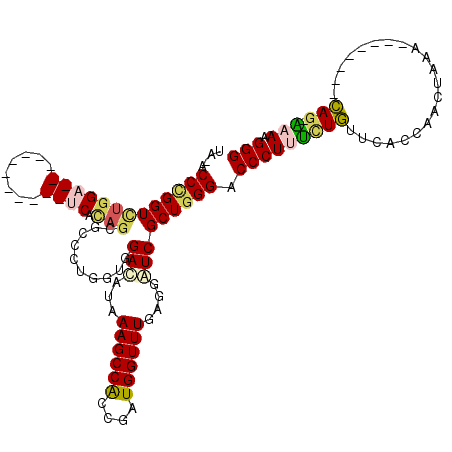
>X_DroMel_CAF1 9600463 100 + 22224390 GUGGAUAUAAAGCCACCGAUGGUUUGAGGAUCGCUGGGACCCUUCCUGUUCACCAACUAAAACAGCGCAAGGGUUCCACACCAUAGAU-ACUACGUACCAC ((((........))))...((((.((((.(((..((((((((((((((((..........))))).).)))))))))).......)))-.)).)).)))). ( -29.40) >DroSec_CAF1 59093 100 + 1 GUGGAUAUAAAGCCACCAAUGGUUUGAGGGUCGCUGGGACCCUUCCUGUUCACCAACUAAAGCAGUGCAAGGGUUCCACACCAUAGAU-ACUACGUACCAC ((((.(((.((((((....))))))...(((.(.((((((((((((((((..........))))).).))))))))))))))......-.....))))))) ( -32.00) >DroSim_CAF1 46304 100 + 1 GUGGAUAUAAAGCCACCAAUGGUUUGAGGGUCGCUGGGACCCUUCCUGUUCACCAACUAAAGCAGUGCAAGGGUUCCACACCAUAGAU-ACUACGUACCAC ((((.(((.((((((....))))))...(((.(.((((((((((((((((..........))))).).))))))))))))))......-.....))))))) ( -32.00) >DroEre_CAF1 63961 100 + 1 GUGGAUAUCAAGCCUCCCAUGGUUUGAGGAUCGCUGGGACCCUUCCUGUUCACCAACUAAAGCAGCGCAAGGGUUCCACACCUCAAAC-ACUACGUACUAC ((((((.(((((((......)))))))..)))(.((((((((((((((((..........))))).).)))))))))))........)-)).......... ( -33.90) >DroYak_CAF1 62726 100 + 1 GUGGAUAUCAAGCCACCCAUGGUUUGAGGAUCGCUGGGACCCUUCCUGUUCACCAACUAAAGCAGCGCAAGGGUUCCACACCAUGAAU-ACCACGUACUAC ((((...((((((((....))))))))((...(.((((((((((((((((..........))))).).))))))))))).))......-.))))....... ( -35.30) >DroMoj_CAF1 84646 99 + 1 GUAGACAUCAAGCCGCCGAUGGUUUGAGGAUCGCUGGGACCCUUUUUGUUUACCAAUUGAA-CAG-AAAUGGGUUCCUGGACGCAGUUCAAUGUGCACCAC ...((..((((((((....))))))))...))...(((((((((((((((((.....))))-)))-))).)))))))(((.((((......))))..))). ( -34.70) >consensus GUGGAUAUAAAGCCACCAAUGGUUUGAGGAUCGCUGGGACCCUUCCUGUUCACCAACUAAAGCAGCGCAAGGGUUCCACACCAUAGAU_ACUACGUACCAC ((((.....(((((......)))))..((.....((((((((((((((((..........))))).).))))))))))..))........))))....... (-24.48 = -24.23 + -0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:55 2006