| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,596,359 – 9,596,454 |
| Length | 95 |
| Max. P | 0.755999 |

| Location | 9,596,359 – 9,596,454 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -24.54 |
| Energy contribution | -25.27 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
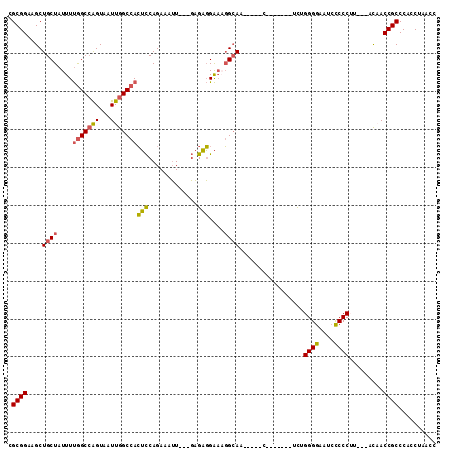
>X_DroMel_CAF1 9596359 95 + 22224390 GGUUAGGUGGGCGGUUGU---AAGGGGUAUUCCCCAAA-------G-----UUGCCUUUCCUCUC---AAUUUCUGGAGUGGCCAAUUACUGGCCAAAAUAGCAGCUUCCGCG ..........((((....---..((((....)))).((-------(-----((((..((((....---.......))))((((((.....)))))).....))))))))))). ( -34.10) >DroSec_CAF1 55022 95 + 1 GGUUAGGUGGGCGGUUGU---AAGGGGGAUUCCCCAGA-------G-----UUGCCUUUCCUCUC---AAUUUCUGGAGUGGCCAAUUACUGGCCAAAAUAGCAGCUUCCGCG ..........((((....---..((((....)))).((-------(-----((((..((((....---.......))))((((((.....)))))).....))))))))))). ( -34.20) >DroSim_CAF1 41522 95 + 1 GGUUAGGUGGGCGGUUGU---AAGGGGGAUUCCCCAGA-------G-----UUGCCUUUCCUCUC---AAUUUCUGGAGUGGCCAAUUACUGGCCAAAAUAGCAGCUUCCGCG ..........((((....---..((((....)))).((-------(-----((((..((((....---.......))))((((((.....)))))).....))))))))))). ( -34.20) >DroEre_CAF1 59830 102 + 1 GGUUGGGUGGGCGGUAGU---ACGGGGGAUUCCCCAGAUACCCCAG-----UUGCCGUUCCUCUC---AAUUUCUGGAGUGGCCAAUUACUGGCCAAAAUAGCAGCUUCCGCG (((((((.(((((((((.---..((((...((....))..))))..-----)))))).))).)))---)))).(((...((((((.....))))))...)))..((....)). ( -37.80) >DroYak_CAF1 58299 107 + 1 GGUUAGGUGGGCGGUAGU---AAGGGAGAUUCCCCAGAUUCGCCAGCUUCGUUGCCUUUCCUCUC---AAUUUCUGGAGUGGCCAAUUACUGGCCAAAAUAGCAGCUUCCGCG ((..((((.(((((....---..(((.....))).....))))).)))).(((((..((((....---.......))))((((((.....)))))).....)))))..))... ( -33.70) >DroAna_CAF1 69059 101 + 1 GGCUUGGUGGGCGGUUAAAGAGCGGGGGAUUCCCCCGC-------G-----AUUCAAGCCAACGGAAUACUCUCUGAUUUGGCCAAUUACCCGCUGAAAUAGCAGCUUCCGCA .(((...(.(((((.(((...(((((((...)))))))-------.-----......(((((((((......))))..)))))...))).))))).)...))).((....)). ( -39.00) >consensus GGUUAGGUGGGCGGUUGU___AAGGGGGAUUCCCCAGA_______G_____UUGCCUUUCCUCUC___AAUUUCUGGAGUGGCCAAUUACUGGCCAAAAUAGCAGCUUCCGCG ......(((((.(((((......((((....))))......................................(((...((((((.....))))))...))))))))))))). (-24.54 = -25.27 + 0.72)


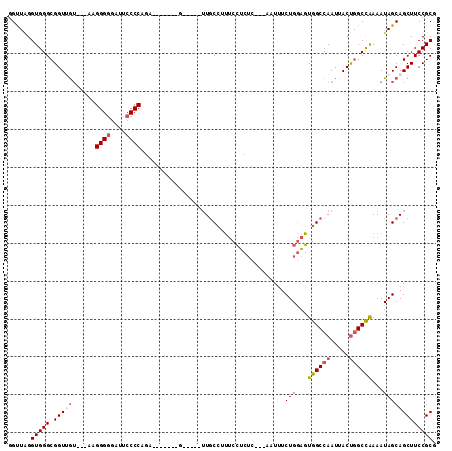
| Location | 9,596,359 – 9,596,454 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -23.00 |
| Energy contribution | -22.87 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
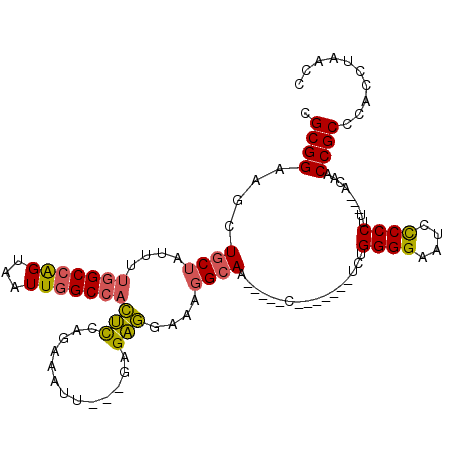
>X_DroMel_CAF1 9596359 95 - 22224390 CGCGGAAGCUGCUAUUUUGGCCAGUAAUUGGCCACUCCAGAAAUU---GAGAGGAAAGGCAA-----C-------UUUGGGGAAUACCCCUU---ACAACCGCCCACCUAACC .(((((((.((((.((((((((((...)))))).(((.(....).---))).)))).)))).-----)-------)).((((....))))..---....)))).......... ( -30.90) >DroSec_CAF1 55022 95 - 1 CGCGGAAGCUGCUAUUUUGGCCAGUAAUUGGCCACUCCAGAAAUU---GAGAGGAAAGGCAA-----C-------UCUGGGGAAUCCCCCUU---ACAACCGCCCACCUAACC .((((.((.((((.((((((((((...)))))).(((.(....).---))).)))).)))).-----)-------)..((((....))))..---....)))).......... ( -30.30) >DroSim_CAF1 41522 95 - 1 CGCGGAAGCUGCUAUUUUGGCCAGUAAUUGGCCACUCCAGAAAUU---GAGAGGAAAGGCAA-----C-------UCUGGGGAAUCCCCCUU---ACAACCGCCCACCUAACC .((((.((.((((.((((((((((...)))))).(((.(....).---))).)))).)))).-----)-------)..((((....))))..---....)))).......... ( -30.30) >DroEre_CAF1 59830 102 - 1 CGCGGAAGCUGCUAUUUUGGCCAGUAAUUGGCCACUCCAGAAAUU---GAGAGGAACGGCAA-----CUGGGGUAUCUGGGGAAUCCCCCGU---ACUACCGCCCACCCAACC .((((.((.(((..((((((((((...)))))).(((.(....).---))).))))..))).-----))..(((((..((((....))))))---))).)))).......... ( -34.50) >DroYak_CAF1 58299 107 - 1 CGCGGAAGCUGCUAUUUUGGCCAGUAAUUGGCCACUCCAGAAAUU---GAGAGGAAAGGCAACGAAGCUGGCGAAUCUGGGGAAUCUCCCUU---ACUACCGCCCACCUAACC .((....))........(((((((...)))))))(((.(....).---)))(((...(((......)))((((.....((((.....)))).---.....))))..))).... ( -32.90) >DroAna_CAF1 69059 101 - 1 UGCGGAAGCUGCUAUUUCAGCGGGUAAUUGGCCAAAUCAGAGAGUAUUCCGUUGGCUUGAAU-----C-------GCGGGGGAAUCCCCCGCUCUUUAACCGCCCACCAAGCC ...((..((((......))))((((....((((((....(((....)))..))))))((((.-----.-------(((((((...)))))))..))))...)))).))..... ( -35.30) >consensus CGCGGAAGCUGCUAUUUUGGCCAGUAAUUGGCCACUCCAGAAAUU___GAGAGGAAAGGCAA_____C_______UCUGGGGAAUCCCCCUU___ACAACCGCCCACCUAACC .((((....((((....(((((((...)))))))(((.............)))....)))).................((((....)))).........)))).......... (-23.00 = -22.87 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:50 2006