
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,595,187 – 9,595,299 |
| Length | 112 |
| Max. P | 0.999476 |

| Location | 9,595,187 – 9,595,299 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -41.81 |
| Consensus MFE | -34.26 |
| Energy contribution | -34.58 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.997849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9595187 112 + 22224390 AGAUGGCCAUAGCGGGUGGAAGAGAGAUGG-UAGAU------CGGGGUCGGGAGCUGAGGUAGAUAAAAGGUGAGGAACCUGCAGCGCCAUUUGCUGAUAGCGCCCGCUGUGAAUUCGA- .((....(((((((((((..((..((((((-(.(((------....)))....((((((((................)))).))))))))))).)).....)))))))))))...))..- ( -39.49) >DroSec_CAF1 53857 112 + 1 AGAUGGCCAUAGCGGGUGCAAGCGAGAUGG-UAGAU------CGGGGUCGGGAGCUGAGAUAGAUAAAAGGUGAGGAACCUGCAGCGCCAUUUGCUGAUAGCGCCCGCUGUGAAUUCGU- .((....((((((((((((.(((.((((((-(.(((------....)))....((((...........((((.....)))).))))))))))))))....))))))))))))...))..- ( -46.10) >DroSim_CAF1 40350 112 + 1 AGAUGGCCAUAGCGGGUGCAAGCGAGAUGG-UAGAU------CGGGGUCGGGAGCUGAGGUAGAUAAAAGGUGAGGAACCUGCAGCGCCAUUUGCUGAUAGCGCCCGCUGUGAAUUCGU- .((....((((((((((((.(((.((((((-(.(((------....)))....((((((((................)))).))))))))))))))....))))))))))))...))..- ( -46.59) >DroEre_CAF1 58728 111 + 1 AGAGGCCCAUAGCGGGUGGAAGAGAGAUGG-GAGAU-------CGGGUCGGGAGCUGAGGUAGAUAAAAGGUGAGGAACCUGCAGCGCCAUUUGCUGAUAGCGCCCGCUGUGAGUUCGA- .(((.(.(((((((((((..((..((((((-..(((-------...)))....((((((((................)))).)))).)))))).)).....))))))))))).))))..- ( -37.39) >DroYak_CAF1 57091 120 + 1 AGAGGGCCAUAGCGGGUGGAAGUGAGAUGGGGAGAUAGGAGUCGGGGUCGGGAGCUGAGGUAGAUAAAAGGUGAGGAACCUGCAGCGCCAUUUGCUGAUAGCGCCCGCUGUGAAUUCAAA .(((...(((((((((((..(((.((((((...(((....)))..........((((((((................)))).)))).))))))))).....)))))))))))..)))... ( -39.49) >consensus AGAUGGCCAUAGCGGGUGGAAGAGAGAUGG_UAGAU______CGGGGUCGGGAGCUGAGGUAGAUAAAAGGUGAGGAACCUGCAGCGCCAUUUGCUGAUAGCGCCCGCUGUGAAUUCGA_ .......(((((((((((..((((((.......(((..........))).((.((((...........((((.....)))).)))).)).)))))).....)))))))))))........ (-34.26 = -34.58 + 0.32)


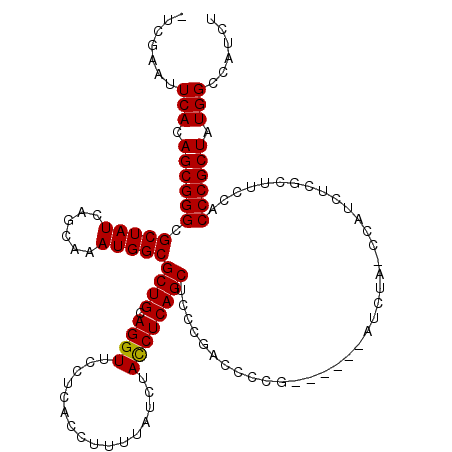
| Location | 9,595,187 – 9,595,299 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -26.07 |
| Energy contribution | -25.91 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9595187 112 - 22224390 -UCGAAUUCACAGCGGGCGCUAUCAGCAAAUGGCGCUGCAGGUUCCUCACCUUUUAUCUACCUCAGCUCCCGACCCCG------AUCUA-CCAUCUCUCUUCCACCCGCUAUGGCCAUCU -......(((.((((((.(((((......)))))((((.((((................))))))))...........------.....-..............)))))).)))...... ( -26.39) >DroSec_CAF1 53857 112 - 1 -ACGAAUUCACAGCGGGCGCUAUCAGCAAAUGGCGCUGCAGGUUCCUCACCUUUUAUCUAUCUCAGCUCCCGACCCCG------AUCUA-CCAUCUCGCUUGCACCCGCUAUGGCCAUCU -......(((.((((((.((....(((....((.((((.((((................)))))))).))((....))------.....-.......))).)).)))))).)))...... ( -28.79) >DroSim_CAF1 40350 112 - 1 -ACGAAUUCACAGCGGGCGCUAUCAGCAAAUGGCGCUGCAGGUUCCUCACCUUUUAUCUACCUCAGCUCCCGACCCCG------AUCUA-CCAUCUCGCUUGCACCCGCUAUGGCCAUCU -......(((.((((((.((....(((....((.((((.((((................)))))))).))((....))------.....-.......))).)).)))))).)))...... ( -31.39) >DroEre_CAF1 58728 111 - 1 -UCGAACUCACAGCGGGCGCUAUCAGCAAAUGGCGCUGCAGGUUCCUCACCUUUUAUCUACCUCAGCUCCCGACCCG-------AUCUC-CCAUCUCUCUUCCACCCGCUAUGGGCCUCU -..((.((((.((((((.(((((......)))))((((.((((................))))))))..........-------.....-..............)))))).))))..)). ( -29.69) >DroYak_CAF1 57091 120 - 1 UUUGAAUUCACAGCGGGCGCUAUCAGCAAAUGGCGCUGCAGGUUCCUCACCUUUUAUCUACCUCAGCUCCCGACCCCGACUCCUAUCUCCCCAUCUCACUUCCACCCGCUAUGGCCCUCU .......(((.((((((.(((((......)))))((((.((((................)))))))).....................................)))))).)))...... ( -26.39) >consensus _UCGAAUUCACAGCGGGCGCUAUCAGCAAAUGGCGCUGCAGGUUCCUCACCUUUUAUCUACCUCAGCUCCCGACCCCG______AUCUA_CCAUCUCGCUUCCACCCGCUAUGGCCAUCU .......(((.((((((.(((((......)))))((((.((((................)))))))).....................................)))))).)))...... (-26.07 = -25.91 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:48 2006