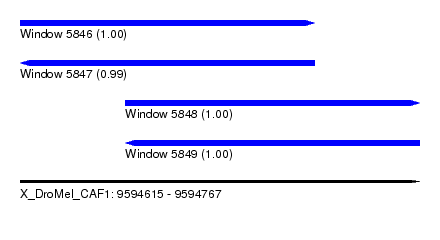
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,594,615 – 9,594,767 |
| Length | 152 |
| Max. P | 0.999994 |

| Location | 9,594,615 – 9,594,727 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -37.04 |
| Energy contribution | -36.28 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.02 |
| SVM RNA-class probability | 0.999759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9594615 112 + 22224390 UCCACCGACUCCAAAUCGAAUCGGAUCGCAAUAGUUAACAAUUUGUUUGGC--------ACUCUGGCACCCUAGGGGGCGCCACUUGGCGCCAGUCGGCGUACCUUCUUCGGUUUGAAAA ...........(((((((((..((..((((((((........)))))((((--------.((((((....))))))((((((....)))))).)))))))..))...))))))))).... ( -37.50) >DroSec_CAF1 53328 112 + 1 UCCACCGACUCCAAAUCGAAUCGGAUCACAAUAGUUAACAAUUUGUUUGGC--------ACUCUGGCACCCCAGGGGGCGCCACUUGGCGCCAGUCGGCGUACCUUCUUCGGGUCGAAAA ....(((((((((((.(((((.(....((....))...).)))))))))).--------.((((((....))))))((((((....))))))))))))((.((((.....)))))).... ( -39.80) >DroSim_CAF1 39821 112 + 1 UCCACCGACUCCAAAUCGAAUCGGAUCACAAUAGUUAACAAUUUGUUUGGC--------ACUCUGGCACCCCAGGGGGCGCCACUUGGCGCCAGUCGGCGUACCUUCUUCGGGUCGAAAA ....(((((((((((.(((((.(....((....))...).)))))))))).--------.((((((....))))))((((((....))))))))))))((.((((.....)))))).... ( -39.80) >DroEre_CAF1 58139 112 + 1 UCCACCGAUUCCAAAUCGAAUCGGAUCACAAUAGUUAACCAUUUGUUUGGC--------ACCCUGGCACCCUAGGGGGCGCCACUUGGCGCCAGUCGGCGUACCUUCUUCGGGUCGAAAA .((.(((((((......)))))))........................(((--------.((((((....))))))((((((....)))))).)))))((.((((.....)))))).... ( -40.60) >DroYak_CAF1 56547 120 + 1 UCCACCGACUCCACAUCGAAUCGGAUCACAAUAGUUAACAAUUUGUUUGGCACUCUGGCACCCUGGCACCCUAGGGGGCGCCACUUGGCGCCAGUCGGCGUACCUUCUUCGGUUCGAGAA ...............((((((((((..((((...........))))..((.((.(((((.((((((....))))))((((((....)))))).))))).)).))...))))))))))... ( -44.80) >consensus UCCACCGACUCCAAAUCGAAUCGGAUCACAAUAGUUAACAAUUUGUUUGGC________ACUCUGGCACCCUAGGGGGCGCCACUUGGCGCCAGUCGGCGUACCUUCUUCGGGUCGAAAA ....(((((((((((.(((((.(....((....))...).))))))))))..........((((((....))))))((((((....))))))))))))((..((......))..)).... (-37.04 = -36.28 + -0.76)

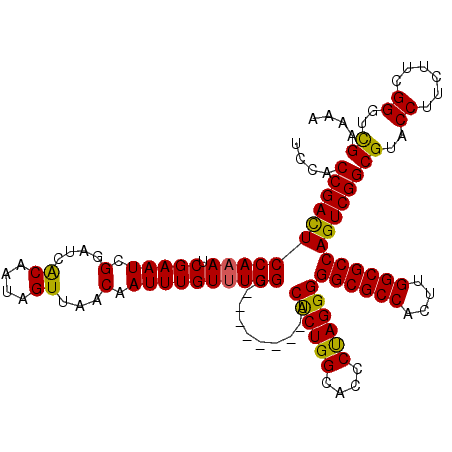

| Location | 9,594,615 – 9,594,727 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -46.90 |
| Consensus MFE | -36.64 |
| Energy contribution | -36.68 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9594615 112 - 22224390 UUUUCAAACCGAAGAAGGUACGCCGACUGGCGCCAAGUGGCGCCCCCUAGGGUGCCAGAGU--------GCCAAACAAAUUGUUAACUAUUGCGAUCCGAUUCGAUUUGGAGUCGGUGGA .......(((......))).(((((((((((((....((((((((....))))))))..))--------))))......((((........))))(((((......)))))))))))).. ( -44.80) >DroSec_CAF1 53328 112 - 1 UUUUCGACCCGAAGAAGGUACGCCGACUGGCGCCAAGUGGCGCCCCCUGGGGUGCCAGAGU--------GCCAAACAAAUUGUUAACUAUUGUGAUCCGAUUCGAUUUGGAGUCGGUGGA ((((((...)))))).....(((((((((((((....((((((((....))))))))..))--------)))...(((((((......((((.....)))).))))))).)))))))).. ( -46.20) >DroSim_CAF1 39821 112 - 1 UUUUCGACCCGAAGAAGGUACGCCGACUGGCGCCAAGUGGCGCCCCCUGGGGUGCCAGAGU--------GCCAAACAAAUUGUUAACUAUUGUGAUCCGAUUCGAUUUGGAGUCGGUGGA ((((((...)))))).....(((((((((((((....((((((((....))))))))..))--------)))...(((((((......((((.....)))).))))))).)))))))).. ( -46.20) >DroEre_CAF1 58139 112 - 1 UUUUCGACCCGAAGAAGGUACGCCGACUGGCGCCAAGUGGCGCCCCCUAGGGUGCCAGGGU--------GCCAAACAAAUGGUUAACUAUUGUGAUCCGAUUCGAUUUGGAAUCGGUGGA ((((((...)))))).....((((((.(((((((...((((((((....)))))))).)))--------)))).((((.(((....)))))))..(((((......))))).)))))).. ( -47.20) >DroYak_CAF1 56547 120 - 1 UUCUCGAACCGAAGAAGGUACGCCGACUGGCGCCAAGUGGCGCCCCCUAGGGUGCCAGGGUGCCAGAGUGCCAAACAAAUUGUUAACUAUUGUGAUCCGAUUCGAUGUGGAGUCGGUGGA ........(((.....(((((.....((((((((...((((((((....)))))))).)))))))).)))))........................(((((((......)))))))))). ( -50.10) >consensus UUUUCGACCCGAAGAAGGUACGCCGACUGGCGCCAAGUGGCGCCCCCUAGGGUGCCAGAGU________GCCAAACAAAUUGUUAACUAUUGUGAUCCGAUUCGAUUUGGAGUCGGUGGA (((((.....))))).(((((.((....((((((....)))))).....)))))))..............((((((.....)))............(((((((......)))))))))). (-36.64 = -36.68 + 0.04)

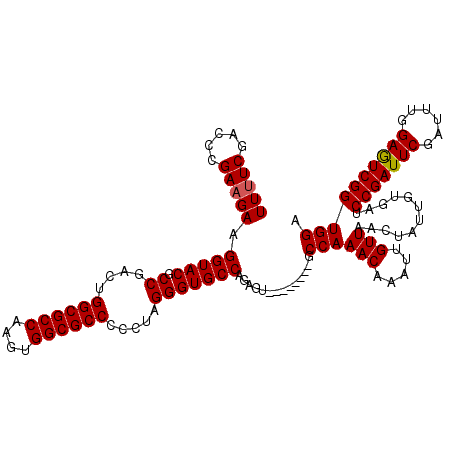

| Location | 9,594,655 – 9,594,767 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.97 |
| Mean single sequence MFE | -48.14 |
| Consensus MFE | -47.28 |
| Energy contribution | -47.04 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.80 |
| SVM RNA-class probability | 0.999994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9594655 112 + 22224390 AUUUGUUUGGC--------ACUCUGGCACCCUAGGGGGCGCCACUUGGCGCCAGUCGGCGUACCUUCUUCGGUUUGAAAACCCAUCGAUCCCAGCGGCAAAUUAGCCACUGCAACGCCCC ........(((--------.((((((....))))))((((((....)))))).)))(((((.........((.((((.......)))).))(((.(((......))).)))..))))).. ( -41.70) >DroSec_CAF1 53368 112 + 1 AUUUGUUUGGC--------ACUCUGGCACCCCAGGGGGCGCCACUUGGCGCCAGUCGGCGUACCUUCUUCGGGUCGAAAACCCAUCGAUCCCAGCGGCAAAUUAGCCACUGCAACGCCCC ........(((--------.((((((....))))))((((((....)))))).)))(((((.........(((((((.......)))))))(((.(((......))).)))..))))).. ( -50.00) >DroSim_CAF1 39861 112 + 1 AUUUGUUUGGC--------ACUCUGGCACCCCAGGGGGCGCCACUUGGCGCCAGUCGGCGUACCUUCUUCGGGUCGAAAACCCAUCGAUCCCAGCGGCAAAUUAGCCACUGCAACGCCCC ........(((--------.((((((....))))))((((((....)))))).)))(((((.........(((((((.......)))))))(((.(((......))).)))..))))).. ( -50.00) >DroEre_CAF1 58179 112 + 1 AUUUGUUUGGC--------ACCCUGGCACCCUAGGGGGCGCCACUUGGCGCCAGUCGGCGUACCUUCUUCGGGUCGAAAACCCAUCGAUCCCAGCGGCAAAUUAGCCACUGCAACGCCCC ........(((--------.((((((....))))))((((((....)))))).)))(((((.........(((((((.......)))))))(((.(((......))).)))..))))).. ( -50.70) >DroYak_CAF1 56587 120 + 1 AUUUGUUUGGCACUCUGGCACCCUGGCACCCUAGGGGGCGCCACUUGGCGCCAGUCGGCGUACCUUCUUCGGUUCGAGAACCCAUCGAUCCCAGCGGCAAAUUAGCCACUGCAACGCCCC ........(((((.(((((.((((((....))))))((((((....)))))).))))).)).........((.((((.......)))).))(((.(((......))).)))....))).. ( -48.30) >consensus AUUUGUUUGGC________ACUCUGGCACCCUAGGGGGCGCCACUUGGCGCCAGUCGGCGUACCUUCUUCGGGUCGAAAACCCAUCGAUCCCAGCGGCAAAUUAGCCACUGCAACGCCCC ........(((.........((((((....))))))((((((....)))))).)))(((((.........(((((((.......)))))))(((.(((......))).)))..))))).. (-47.28 = -47.04 + -0.24)

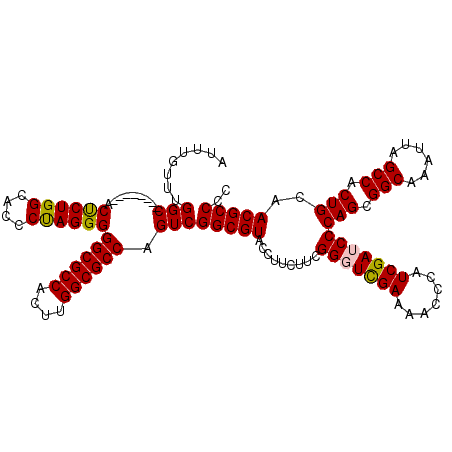
| Location | 9,594,655 – 9,594,767 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.97 |
| Mean single sequence MFE | -55.24 |
| Consensus MFE | -48.32 |
| Energy contribution | -48.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9594655 112 - 22224390 GGGGCGUUGCAGUGGCUAAUUUGCCGCUGGGAUCGAUGGGUUUUCAAACCGAAGAAGGUACGCCGACUGGCGCCAAGUGGCGCCCCCUAGGGUGCCAGAGU--------GCCAAACAAAU ..((((((.(((((((......))))))).))((..(.((((....)))).).)).....))))...((((((....((((((((....))))))))..))--------))))....... ( -48.70) >DroSec_CAF1 53368 112 - 1 GGGGCGUUGCAGUGGCUAAUUUGCCGCUGGGAUCGAUGGGUUUUCGACCCGAAGAAGGUACGCCGACUGGCGCCAAGUGGCGCCCCCUGGGGUGCCAGAGU--------GCCAAACAAAU ..(((((.((((((((......))))))(((.((((.......))))))).......)))))))...((((((....((((((((....))))))))..))--------))))....... ( -54.70) >DroSim_CAF1 39861 112 - 1 GGGGCGUUGCAGUGGCUAAUUUGCCGCUGGGAUCGAUGGGUUUUCGACCCGAAGAAGGUACGCCGACUGGCGCCAAGUGGCGCCCCCUGGGGUGCCAGAGU--------GCCAAACAAAU ..(((((.((((((((......))))))(((.((((.......))))))).......)))))))...((((((....((((((((....))))))))..))--------))))....... ( -54.70) >DroEre_CAF1 58179 112 - 1 GGGGCGUUGCAGUGGCUAAUUUGCCGCUGGGAUCGAUGGGUUUUCGACCCGAAGAAGGUACGCCGACUGGCGCCAAGUGGCGCCCCCUAGGGUGCCAGGGU--------GCCAAACAAAU ..(((((.((((((((......))))))(((.((((.......))))))).......)))))))...(((((((...((((((((....)))))))).)))--------))))....... ( -57.50) >DroYak_CAF1 56587 120 - 1 GGGGCGUUGCAGUGGCUAAUUUGCCGCUGGGAUCGAUGGGUUCUCGAACCGAAGAAGGUACGCCGACUGGCGCCAAGUGGCGCCCCCUAGGGUGCCAGGGUGCCAGAGUGCCAAACAAAU (((((((..(((((((......)))))))((.((((.......)))).)).........)))))..((((((((...((((((((....)))))))).))))))))....))........ ( -60.60) >consensus GGGGCGUUGCAGUGGCUAAUUUGCCGCUGGGAUCGAUGGGUUUUCGACCCGAAGAAGGUACGCCGACUGGCGCCAAGUGGCGCCCCCUAGGGUGCCAGAGU________GCCAAACAAAU (((((((..(((((((......)))))))((.((((.......)))).)).........)))))..((((((((....)))((((....)))))))))............))........ (-48.32 = -48.52 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:46 2006