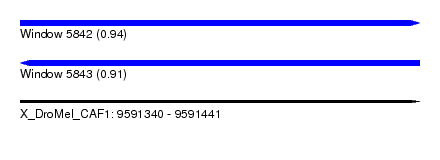
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,591,340 – 9,591,441 |
| Length | 101 |
| Max. P | 0.944683 |

| Location | 9,591,340 – 9,591,441 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -21.44 |
| Consensus MFE | -11.31 |
| Energy contribution | -11.37 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
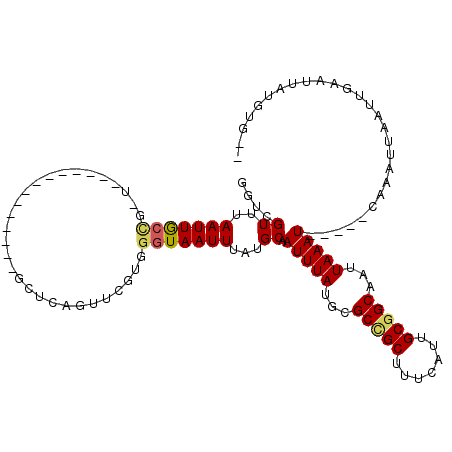
>X_DroMel_CAF1 9591340 101 + 22224390 --CACAUAAUUCAAUUAAUUUG-----AUUUAAUUGCCGCAAUGAAAGCGGCGCAUAAAUUGCAUAAAUUACCCACGAACUGAGCAA---------UGCACCGGCAAUUAAACGACC --........((((.....)))-----)((((((((((((.......))((.((((....(((....................))))---------))).))))))))))))..... ( -22.15) >DroPse_CAF1 66268 100 + 1 CACACAUAAUUCAAUUAAUUUGAUUUGAUUUAAUUGCCGCAAUGAAAGCGGCUCAUAAAUUGCAUAAAUUACCCACGAACUAGUG-----------------AGUAAUUAUACGAGU ........((((..............((((((...(((((.......)))))...)))))).....((((((.(((......)))-----------------.))))))....)))) ( -21.30) >DroSim_CAF1 36567 101 + 1 --CACAUAAUUCAAUUAAUUUG-----AUUUAAUUGCCGCAAUGAAAGCGGCGCAUAAAUUGCAUAAAUUACCCACGAAAUGAGCAA---------UGCACCGGCAAUUAAACGACC --........((((.....)))-----)((((((((((((.......))((.((((....(((....(((........)))..))))---------))).))))))))))))..... ( -22.20) >DroYak_CAF1 52764 110 + 1 --CACAUAAUUCAAUUAAUUUG-----AUUUAAUUGCCGCAAUGAAAGCAGCGCAUAAAUUGCAUAAAUUACCCACGAACUGAGCAACUGAGCAAUUGCAGCGGCAAUUAAACGACC --........((((.....)))-----)((((((((((((............(((.....)))....................((((........)))).))))))))))))..... ( -25.20) >DroAna_CAF1 64348 81 + 1 --CACAUAAUUCAAUUAAUUUG-----AUUUAAUUGCUGCAAUGAAAGCGGCGCAUAAAUUGCAUAAAUUA----------------------------AAAGGCAAUUAAACG-CA --................((((-----(((((...(((((.......)))))(((.....))).)))))))----------------------------))..((........)-). ( -16.50) >DroPer_CAF1 67115 100 + 1 CACACAUAAUUCAAUUAAUUUGAUUUGAUUUAAUUGCCGCAAUGAAAGCGGCUCAUAAAUUGCAUAAAUUACCCACGAACUAGUG-----------------AGUAAUUAUACGAGU ........((((..............((((((...(((((.......)))))...)))))).....((((((.(((......)))-----------------.))))))....)))) ( -21.30) >consensus __CACAUAAUUCAAUUAAUUUG_____AUUUAAUUGCCGCAAUGAAAGCGGCGCAUAAAUUGCAUAAAUUACCCACGAACUGAGC______________A_CGGCAAUUAAACGACC ...............(((((((.....(((((...(((((.......)))))...)))))....))))))).............................................. (-11.31 = -11.37 + 0.06)



| Location | 9,591,340 – 9,591,441 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.69 |
| Mean single sequence MFE | -25.94 |
| Consensus MFE | -12.77 |
| Energy contribution | -12.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9591340 101 - 22224390 GGUCGUUUAAUUGCCGGUGCA---------UUGCUCAGUUCGUGGGUAAUUUAUGCAAUUUAUGCGCCGCUUUCAUUGCGGCAAUUAAAU-----CAAAUUAAUUGAAUUAUGUG-- ....(((((((((((((((((---------(((((((.....)))))).............)))))))((.......)))))))))))))-----....................-- ( -28.41) >DroPse_CAF1 66268 100 - 1 ACUCGUAUAAUUACU-----------------CACUAGUUCGUGGGUAAUUUAUGCAAUUUAUGAGCCGCUUUCAUUGCGGCAAUUAAAUCAAAUCAAAUUAAUUGAAUUAUGUGUG ....(((((((((((-----------------(((......)))))))).))))))....((((((((((.......)))))............((((.....)))).))))).... ( -26.30) >DroSim_CAF1 36567 101 - 1 GGUCGUUUAAUUGCCGGUGCA---------UUGCUCAUUUCGUGGGUAAUUUAUGCAAUUUAUGCGCCGCUUUCAUUGCGGCAAUUAAAU-----CAAAUUAAUUGAAUUAUGUG-- ....(((((((((((((((((---------((((((((...))))))).............)))))))((.......)))))))))))))-----....................-- ( -29.01) >DroYak_CAF1 52764 110 - 1 GGUCGUUUAAUUGCCGCUGCAAUUGCUCAGUUGCUCAGUUCGUGGGUAAUUUAUGCAAUUUAUGCGCUGCUUUCAUUGCGGCAAUUAAAU-----CAAAUUAAUUGAAUUAUGUG-- ....(((((((((((((.((((((((..(((((((((.....)))))))))...)))))...))))).((.......)))))))))))))-----....................-- ( -31.40) >DroAna_CAF1 64348 81 - 1 UG-CGUUUAAUUGCCUUU----------------------------UAAUUUAUGCAAUUUAUGCGCCGCUUUCAUUGCAGCAAUUAAAU-----CAAAUUAAUUGAAUUAUGUG-- .(-(((..((((((....----------------------------........))))))...)))).((.......))..(((((((..-----....))))))).........-- ( -14.20) >DroPer_CAF1 67115 100 - 1 ACUCGUAUAAUUACU-----------------CACUAGUUCGUGGGUAAUUUAUGCAAUUUAUGAGCCGCUUUCAUUGCGGCAAUUAAAUCAAAUCAAAUUAAUUGAAUUAUGUGUG ....(((((((((((-----------------(((......)))))))).))))))....((((((((((.......)))))............((((.....)))).))))).... ( -26.30) >consensus GGUCGUUUAAUUGCCG_U______________GCUCAGUUCGUGGGUAAUUUAUGCAAUUUAUGCGCCGCUUUCAUUGCGGCAAUUAAAU_____CAAAUUAAUUGAAUUAUGUG__ ....((..(((((((.............................)))))))...)).(((((...(((((.......)))))...)))))........................... (-12.77 = -12.77 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:41 2006