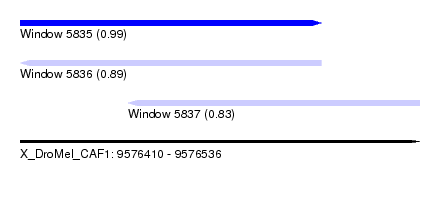
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,576,410 – 9,576,536 |
| Length | 126 |
| Max. P | 0.990049 |

| Location | 9,576,410 – 9,576,505 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.96 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.98 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.990049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9576410 95 + 22224390 AGUUUAGCCCGAAAGUAUCUCAUAGAUACAUAUGUGGGCUCACUGCGCACAAGUGACUGGCAGGCAUUUGCUGACUUAGUCAUGAUUUGAUG--AAU .(((((...((((.(((((.....)))))...((((.((.....)).)))).((((((((..(((....)))...))))))))..)))).))--))) ( -26.10) >DroSec_CAF1 35463 94 + 1 AAUUUAGCCCGA-AGUAUCUCAUAGAUACAUAUGUGGGCUCACUGCGGACAAGUGACUGGCAGGCAUUUGCCGACUUAGUCAUGAUUUGAUG--AAU ......(((((.-.(((((.....))))).....)))))(((...((((...((((((((..(((....)))..).)))))))..)))).))--).. ( -28.60) >DroSim_CAF1 21927 94 + 1 AAUUUAGCCCGA-AGUAUCUCAUAGAUACAUAUGUGGGCUCACUGCGGACAAGUGACUGGCAGGCAUUUGCCGACUUAGUCAUGAUUUGAUG--AAU ......(((((.-.(((((.....))))).....)))))(((...((((...((((((((..(((....)))..).)))))))..)))).))--).. ( -28.60) >DroEre_CAF1 40266 94 + 1 AAUUUAGCCCGA-AGUAUCUCAUAGAUACAUAUGCGAGCGCACUGCAGACAAGUGACUGGCAGGCAUUUGCCGACUAAGUCAUGAUUUGAUG--AAU .(((((((.((.-.(((((.....))))).....)).))......((((...((((((((..(((....)))..)).))))))..)))).))--))) ( -23.50) >DroYak_CAF1 37270 96 + 1 AAUUUAGCCCGA-AGUAUCUCAUAGAUACAUAUGUGAGCUCACUGCAGACAAGUGACUGGCAGGCAUUUGCCGACUUAGUCAUGAUUUGAUGAGAAU ............-.(((((.....))))).........((((...((((...((((((((..(((....)))..).)))))))..)))).))))... ( -24.80) >consensus AAUUUAGCCCGA_AGUAUCUCAUAGAUACAUAUGUGGGCUCACUGCGGACAAGUGACUGGCAGGCAUUUGCCGACUUAGUCAUGAUUUGAUG__AAU ......(((((...(((((.....))))).....)))))......((((...((((((((..(((....)))..)).))))))..))))........ (-24.94 = -24.98 + 0.04)



| Location | 9,576,410 – 9,576,505 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 94.96 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -20.68 |
| Energy contribution | -20.68 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9576410 95 - 22224390 AUU--CAUCAAAUCAUGACUAAGUCAGCAAAUGCCUGCCAGUCACUUGUGCGCAGUGAGCCCACAUAUGUAUCUAUGAGAUACUUUCGGGCUAAACU ...--..........((((...))))((....))((((((........)).))))..(((((......(((((.....)))))....)))))..... ( -19.90) >DroSec_CAF1 35463 94 - 1 AUU--CAUCAAAUCAUGACUAAGUCGGCAAAUGCCUGCCAGUCACUUGUCCGCAGUGAGCCCACAUAUGUAUCUAUGAGAUACU-UCGGGCUAAAUU ...--..........(((((..((.(((....))).)).)))))...(((((..(((....)))....(((((.....))))).-.)))))...... ( -23.50) >DroSim_CAF1 21927 94 - 1 AUU--CAUCAAAUCAUGACUAAGUCGGCAAAUGCCUGCCAGUCACUUGUCCGCAGUGAGCCCACAUAUGUAUCUAUGAGAUACU-UCGGGCUAAAUU ...--..........(((((..((.(((....))).)).)))))...(((((..(((....)))....(((((.....))))).-.)))))...... ( -23.50) >DroEre_CAF1 40266 94 - 1 AUU--CAUCAAAUCAUGACUUAGUCGGCAAAUGCCUGCCAGUCACUUGUCUGCAGUGCGCUCGCAUAUGUAUCUAUGAGAUACU-UCGGGCUAAAUU ...--..........(((((..((.(((....))).)).)))))...(((((..((((....))))..(((((.....))))).-.)))))...... ( -22.80) >DroYak_CAF1 37270 96 - 1 AUUCUCAUCAAAUCAUGACUAAGUCGGCAAAUGCCUGCCAGUCACUUGUCUGCAGUGAGCUCACAUAUGUAUCUAUGAGAUACU-UCGGGCUAAAUU ...............(((((..((.(((....))).)).)))))...(((((..(((....)))....(((((.....))))).-.)))))...... ( -21.90) >consensus AUU__CAUCAAAUCAUGACUAAGUCGGCAAAUGCCUGCCAGUCACUUGUCCGCAGUGAGCCCACAUAUGUAUCUAUGAGAUACU_UCGGGCUAAAUU ...............(((((..((.(((....))).)).)))))...(((((..(((....)))....(((((.....)))))...)))))...... (-20.68 = -20.68 + -0.00)

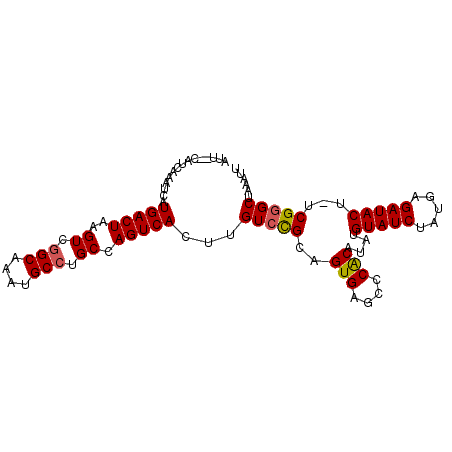

| Location | 9,576,444 – 9,576,536 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 90.04 |
| Mean single sequence MFE | -32.01 |
| Consensus MFE | -26.12 |
| Energy contribution | -25.48 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.831015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9576444 92 - 22224390 AUGUCGAAGUGCACGAGUGUCUG---------CGGGAGGCAUU--CAUCAAAUCAUGACUAAGUCAGCAAAUGCCUGCCAGUCACUUGUGCGCAGUGAGCCCA ...(((..(((((((((((.((.---------.((.(((((((--..........((((...))))...))))))).)))).)))))))))))..)))..... ( -35.82) >DroSec_CAF1 35496 92 - 1 AUGUCGAAGUGCACGAGUGUCUG---------CGGGAGGCAUU--CAUCAAAUCAUGACUAAGUCGGCAAAUGCCUGCCAGUCACUUGUCCGCAGUGAGCCCA ...(((..(((.(((((((.((.---------.((.(((((((--..........((((...))))...))))))).)))).))))))).)))..)))..... ( -28.52) >DroSim_CAF1 21960 92 - 1 AUGUCGAAGUGCACGAGUGUCUG---------CGGGAGGCAUU--CAUCAAAUCAUGACUAAGUCGGCAAAUGCCUGCCAGUCACUUGUCCGCAGUGAGCCCA ...(((..(((.(((((((.((.---------.((.(((((((--..........((((...))))...))))))).)))).))))))).)))..)))..... ( -28.52) >DroEre_CAF1 40299 92 - 1 AUGCCGAGGUGAACGAGUGUCUG---------UGGGAGGCAUU--CAUCAAAUCAUGACUUAGUCGGCAAAUGCCUGCCAGUCACUUGUCUGCAGUGCGCUCG .((((((..(((..((((((((.---------....)))))))--).)))..))..(((...)))))))...((((((.((.(....).)))))).))..... ( -30.20) >DroYak_CAF1 37303 103 - 1 AUGCCGAAGUGCACGAGUGUCUGGCAUUUGAGUGGGAGGCAUUCUCAUCAAAUCAUGACUAAGUCGGCAAAUGCCUGCCAGUCACUUGUCUGCAGUGAGCUCA ..((...(.((((((((((.(((((((((((.((((((...)))))))))))....(((...)))(((....))))))))).)))))))..))).)..))... ( -37.00) >consensus AUGUCGAAGUGCACGAGUGUCUG_________CGGGAGGCAUU__CAUCAAAUCAUGACUAAGUCGGCAAAUGCCUGCCAGUCACUUGUCCGCAGUGAGCCCA ..((((..(((.(((((((.((...........((.(((((((............((((...))))...))))))).)))).))))))).)))..)).))... (-26.12 = -25.48 + -0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:35 2006