
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,568,899 – 9,569,033 |
| Length | 134 |
| Max. P | 0.985525 |

| Location | 9,568,899 – 9,569,012 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 87.21 |
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -35.13 |
| Energy contribution | -34.88 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
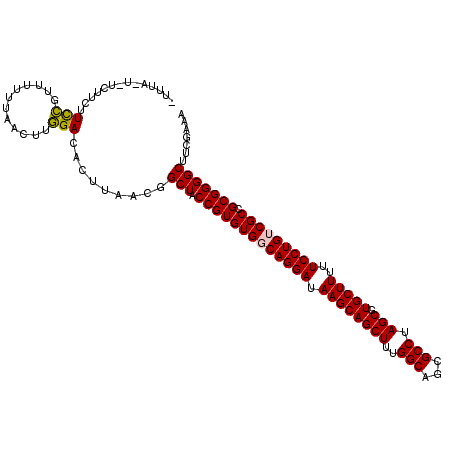
>X_DroMel_CAF1 9568899 113 + 22224390 UUUUGUG-UCUUCUUCCGUUUUUUAACUUGGACACUUAACGGCUACCGUGUGGCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCCGCGGGGCUUCGAAA ....(((-(((......(((....)))..)))))).....((((.(((((((((((((.((((((((..(((...))).))).)))))..)))))))).)))))))))...... ( -43.60) >DroSec_CAF1 28801 110 + 1 -UUUA-U-UCUUCUUCCGUUU-UUAACUUGGACACUUAACGGCUACCGUGUGGCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCCGCGGGGCUUCGAAA -....-(-((....((((...-......))))........((((.(((((((((((((.((((((((..(((...))).))).)))))..)))))))).)))))))))..))). ( -40.20) >DroSim_CAF1 15219 111 + 1 -UUUA-U-UCUUCUUCCGUUUUUUAACUUGGACACUUAACGGCUACCGUGUGGCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCCGCGGGGCUUCGAAA -....-(-((....((((..........))))........((((.(((((((((((((.((((((((..(((...))).))).)))))..)))))))).)))))))))..))). ( -40.10) >DroEre_CAF1 29589 114 + 1 UCUUAUUUUUUCUUUCCUUUUUUUAACUUGGACACUUAACGGCUACCGUGUGGCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCCGCGGGGCUUCGACA ..............(((............)))........((((.(((((((((((((.((((((((..(((...))).))).)))))..)))))))).)))))))))...... ( -38.60) >DroWil_CAF1 48741 109 + 1 ---UAUAUUUGG--UUUUUGUUUUAGCUUAGACACUUAACGGCUACCGUGUGUCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCUGCGGGGCAUUCAAG ---.....((((--((..(((((......)))))...))).(((.(((((((.(((((.((((((((..(((...))).))).)))))..))))).))).)))))))...))). ( -37.50) >DroPer_CAF1 42083 109 + 1 CUCUG---UCGC--UCUUUUCUUUGACUUAGACACUUAACGGCUACCGUGUGUCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCCGCGGGGCAUCGCGA .....---((((--..........(((...(((((...((((...)))))))))((((.((((((((..(((...))).))).)))))..)))))))(((....)))...)))) ( -38.60) >consensus _UUUA_U_UCUUCUUCCGUUUUUUAACUUGGACACUUAACGGCUACCGUGUGGCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCCGCGGGGCUUCGAAA ..............(((............))).........(((.(((((((((((((.((((((((..(((...))).))).)))))..))))))))).)))))))....... (-35.13 = -34.88 + -0.25)


| Location | 9,568,934 – 9,569,033 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -35.10 |
| Energy contribution | -35.43 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9568934 99 + 22224390 UAACGGCUACCGUGUGGCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCCGCGGGGCUUCGA-AAAAAAUAUAGGAUACAUUGGG----G---------- ....((((.(((((((((((((.((((((((..(((...))).))).)))))..)))))))).)))))))))....-......................----.---------- ( -37.40) >DroPse_CAF1 41201 84 + 1 UAACGGCUACCGUGUGUCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCCGCGGGGCAUCGC-GA---------------CUGGG----G---------- .....(((.(((((((.(((((.((((((((..(((...))).))).)))))..))))).)).)))))))).....-..---------------.....----.---------- ( -33.80) >DroSec_CAF1 28833 98 + 1 UAACGGCUACCGUGUGGCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCCGCGGGGCUUCGA-AAAAAAUAUAGGAUACAUGG-G----G---------- ....((((.(((((((((((((.((((((((..(((...))).))).)))))..)))))))).)))))))))....-....................-.----.---------- ( -37.40) >DroEre_CAF1 29625 99 + 1 UAACGGCUACCGUGUGGCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCCGCGGGGCUUCGA-CAAAAAUAUAGGAUGCAUGGGG----G---------- ....((((.(((((((((((((.((((((((..(((...))).))).)))))..)))))))).)))))))))....-......................----.---------- ( -37.40) >DroYak_CAF1 30526 114 + 1 UAACGGCUACCGUGUGGCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCCGCGGGGCUUCGAACUAAAAUAUAGGAUACAUGGGGAUGGGGCGGGGGGUG .....(((.(((((((((((((.((((((((..(((...))).))).)))))..)))))))).)))))..(((((..(((...(((.(....))))....)))..)))))))). ( -42.40) >DroPer_CAF1 42114 84 + 1 UAACGGCUACCGUGUGUCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCCGCGGGGCAUCGC-GA---------------CUGGG----G---------- .....(((.(((((((.(((((.((((((((..(((...))).))).)))))..))))).)).)))))))).....-..---------------.....----.---------- ( -33.80) >consensus UAACGGCUACCGUGUGGCAGGAUAAGCAGCUUUGGCAGCGCCUAGCGUGCUUUUUCCUGUCGCCGCGGGGCUUCGA_AAAAAAUAUAGGAUACAUGGGG____G__________ .....(((.(((((((((((((.((((((((..(((...))).))).)))))..)))))))).))))))))........................................... (-35.10 = -35.43 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:32 2006