
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,553,473 – 9,553,573 |
| Length | 100 |
| Max. P | 0.798090 |

| Location | 9,553,473 – 9,553,573 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.69 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -26.53 |
| Energy contribution | -26.56 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9553473 100 + 22224390 GU-UGGGGCGUGCGCAUUGAUUGAAUGCUCAAUUGAGCGCAUGAACCGGCGAUCAUUUUGUAC-AGCGCCCUCCCCCAGUACCGCCUCCUCCGCCCACCACA----- ((-.(((((((((((.(..(((((....)))))..))))))).....((((........((((-..............))))))))......)))).).)).----- ( -31.74) >DroPse_CAF1 26087 95 + 1 GU-UGGGGCGUGCGCAUUGAUUGAAUGCUCAAUUGAGCGCAUGAACCGGCGAUCAUUUUGCCUGUACGCCCUCCUA---------CUCCUCC-CCCAACG-AGCCAC ((-((((((((((((.(..(((((....)))))..)))))))).((.(((((.....))))).))...........---------......)-)))))).-...... ( -38.10) >DroEre_CAF1 13120 99 + 1 GU-UGGGGCGUGCGCAUUGAUUGAAUGCUCAAUUGAGCGCAUGAACCGGCGAUCAUUUUGUAC-AGCGCCCUCCCCCAGCACCGCCCCCUCCGCCCACCG-A----- ((-((((((((((((.(..(((((....)))))..))))))))....((((............-..))))...)))))))....................-.----- ( -35.54) >DroWil_CAF1 21916 91 + 1 GUUUGGGGCGUGCGCAUUGAUUGAAUGCUCAAUUGAGCGCAUGAACCGGCGAUCAUUUUGUUC-AAUGCCCGACC----------CUCCUCCAGCCACCGCA----- (.(((((((((((((.(..(((((....)))))..))))))))....((((..((((......-))))..)).))----------...)))))).)......----- ( -28.50) >DroYak_CAF1 13711 99 + 1 GU-UGGGGCGUGCGCAUUGAUUGAAUGCUCAAUUGAGCGCAUGAACCGGCGAUCAUUUUGUAC-AGCGACCUCCCCCAGCACCGCCCCCUCCGCCCACCG-A----- ((-((((((((((((.(..(((((....)))))..)))))))).....((((.....))))..-.........)))))))....................-.----- ( -32.20) >DroPer_CAF1 26735 95 + 1 GU-UGGGGCGUGCGCAUUGAUUGAAUGCUCAAUUGAGCGCAUGAACCGGCGAUCAUUUUGCCUGUACGCCCUCCUA---------CUCCUCC-CCCAACG-AGCCAC ((-((((((((((((.(..(((((....)))))..)))))))).((.(((((.....))))).))...........---------......)-)))))).-...... ( -38.10) >consensus GU_UGGGGCGUGCGCAUUGAUUGAAUGCUCAAUUGAGCGCAUGAACCGGCGAUCAUUUUGUAC_AACGCCCUCCCC_________CUCCUCCGCCCACCG_A_____ ....((..(((((((.(..(((((....)))))..))))))))..))((((...............))))..................................... (-26.53 = -26.56 + 0.03)

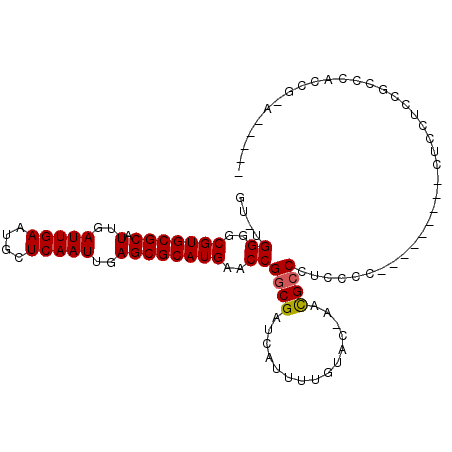
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:29 2006