| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,551,131 – 9,551,228 |
| Length | 97 |
| Max. P | 0.898776 |

| Location | 9,551,131 – 9,551,228 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.64 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9551131 97 - 22224390 --------------UCC-UUU-CUCUCUAAUGCUAAUGCGAG-U-----AUGCAGGCAAUUGGAAAAUUUAUUCAAUUGCUGGCAAUUCAACUAAAUCACUCAUUGGCUUUAUUAU-GUG --------------...-...-.....(((.(((((((.(((-(-----.(((.((((((((((.......)))))))))).)))))))............))))))).)))....-... ( -21.82) >DroGri_CAF1 12304 95 - 1 --------------GCA---U-CCUUCUAAUGCUAAUACGAA-A-----ACGCAGGCAAUUGGAAAAUUUAUUCAAUUGUUGGCAAUUCGACUAAAUCACUCAUUGGCAUUAUUAU-GUG --------------(((---(-.....((((((((((..(((-.-----..((..(((((((((.......)))))))))..))..)))((.....))....))))))))))..))-)). ( -22.00) >DroEre_CAF1 10815 98 - 1 --------------UUCCUCC-CCCACAAAUGCUAAUGCGAG-A-----AUGCAGGCAAUUGGAAAAUUUAUUCAAUUGCUGGCAAUUCAACUAAAUCACUCAUUGGCUUUAUUAU-GUG --------------.......-..((((...(((((((.(((-(-----((((.((((((((((.......)))))))))).)).)))).......))...))))))).......)-))) ( -22.21) >DroWil_CAF1 18070 110 - 1 -------GAGAGAGUCC-UCCGCGCUCUAAUGCUAAUGCGAG-CAAAAGAUUCAGGCAAUUGGAAAAUUUAUUCAAUUGUUGGCAACUCAACUAAAUCGCUCAUUGGCUUUAUUAU-GUG -------.((((.((..-...)).))))...((((((..(((-(....((((.(((((((((((.......))))))))).(....)....)).))))))))))))))........-... ( -24.30) >DroMoj_CAF1 17903 106 - 1 UUGUAUAAGCAGAGGCA---U-CCUUCUAAUGCUAAUACGAA-A-----ACGCAGGCAAUUGGAAAAUUUAUUCAAUUGUUGGCAAUUCAACUAAAUCCCUCAUUGGCUU---UAU-GUG ((((((.((((((((..---.-.))))...)))).)))))).-.-----..((..(((((((((.......)))))))))..))......((((((.((......)).))---)).-)). ( -23.90) >DroAna_CAF1 11398 90 - 1 -------------------------ACUAAUGCUAAUGCGAAAA-----ACGCAGGCAAUUGGAAAAUUUAUUCAAUUGCUGACAAUUCAACUAAAUCACUCAUUGGCUUUAUUAUCCUG -------------------------..(((.(((((((((....-----.))).((((((((((.......)))))))))).....................)))))).)))........ ( -19.00) >consensus ______________UCC___U_CCCUCUAAUGCUAAUGCGAA_A_____ACGCAGGCAAUUGGAAAAUUUAUUCAAUUGCUGGCAAUUCAACUAAAUCACUCAUUGGCUUUAUUAU_GUG ...............................(((((((.((..........((.((((((((((.......)))))))))).))............))...)))))))............ (-15.32 = -15.73 + 0.42)

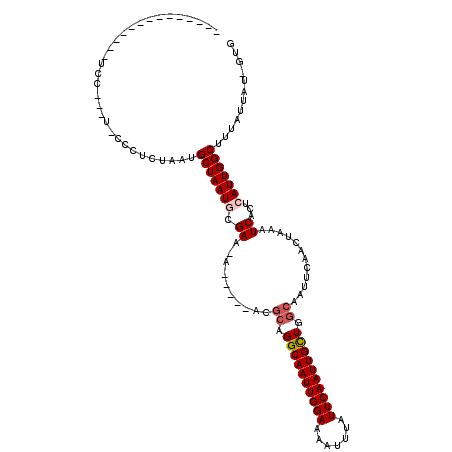

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:28 2006