
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,529,532 – 9,529,670 |
| Length | 138 |
| Max. P | 0.997956 |

| Location | 9,529,532 – 9,529,640 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 93.20 |
| Mean single sequence MFE | -37.46 |
| Consensus MFE | -30.37 |
| Energy contribution | -30.25 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9529532 108 + 22224390 ACUAAAAUGCGACGAGACGUGCCGAAGACGGG-GCGAAAAG-GGAAGA--ACAACGAGAACAG-UACCGAGAAUUCUCUCGGUCUCGUUGCACUCAUCUGUGUGCCGCUUGAC ............((((.(...(((....)))(-(((...((-(..((.--.((((((((....-..((((((....))))))))))))))..))..)))...))))))))).. ( -35.20) >DroSec_CAF1 13367 110 + 1 ACUAAAAUGCGACGAGACGUGCCGAAAACGGG-GCGAAAAG-GGAAGAGUACAACGAGAACGG-UACCGAGAAUUCUCUCGGUCUCGUUGCACUCAUCUGUGUGCCGCUUGAC ............((((.(...(((....)))(-(((...(.-(((.((((.((((((((....-..((((((....)))))))))))))).)))).))).).))))))))).. ( -40.50) >DroSim_CAF1 12870 110 + 1 ACUAAAAUGCGACGAGACGUGCCGAAAACGGG-GCGAAAAG-GGAAGAGUACAACGAGAACGG-UACCGAGAAUUCUCUCGGUCUCGUUGCACUCAUCUGUGUGCCGCUUGAC ............((((.(...(((....)))(-(((...(.-(((.((((.((((((((....-..((((((....)))))))))))))).)))).))).).))))))))).. ( -40.50) >DroEre_CAF1 13675 113 + 1 ACUAAAAUGCGACGAGACGUGCCGAAAACGGGGACGAAAAGGGGAAGAGAACAACGAGAACGGGAAUCGAGAAUUCUCUCGGUCUUGUUGCACUCAUCUGUGCGCCGCUUGAC ............((((..(((((.....((....))......(((.(((..((((((((.(((((...........))))).))))))))..))).)))).))))..)))).. ( -34.30) >DroYak_CAF1 13945 112 + 1 ACUAAAAUGCGACGAGACGUGCCGAAAACGGGGGCGAAAAG-GGAACAGAACAACGAGAACGGGAACCGAGAAUUCUCUCGGUCUCGCUGUACUCUUCUGUGCGCCGCUUGAC ............((((.(((.((.......)).))).....-((.((((((....(((.(((((((((((((....))))))).)).)))).)))))))))...)).)))).. ( -36.80) >consensus ACUAAAAUGCGACGAGACGUGCCGAAAACGGG_GCGAAAAG_GGAAGAGAACAACGAGAACGG_UACCGAGAAUUCUCUCGGUCUCGUUGCACUCAUCUGUGUGCCGCUUGAC ........(((.((...(((.((.......)).)))......(((.(((..((((((((.......((((((....))))))))))))))..))).)))...)).)))..... (-30.37 = -30.25 + -0.12)



| Location | 9,529,532 – 9,529,640 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 93.20 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -32.61 |
| Energy contribution | -33.09 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9529532 108 - 22224390 GUCAAGCGGCACACAGAUGAGUGCAACGAGACCGAGAGAAUUCUCGGUA-CUGUUCUCGUUGU--UCUUCC-CUUUUCGC-CCCGUCUUCGGCACGUCUCGUCGCAUUUUAGU .....(((((....(((((((.((((((((((((((......)))))..-....)))))))))--.))...-......((-(........))).))))).)))))........ ( -36.00) >DroSec_CAF1 13367 110 - 1 GUCAAGCGGCACACAGAUGAGUGCAACGAGACCGAGAGAAUUCUCGGUA-CCGUUCUCGUUGUACUCUUCC-CUUUUCGC-CCCGUUUUCGGCACGUCUCGUCGCAUUUUAGU .....(((((.....((.((((((((((((((((((......)))))..-....))))))))))))).)).-......((-(........))).......)))))........ ( -42.10) >DroSim_CAF1 12870 110 - 1 GUCAAGCGGCACACAGAUGAGUGCAACGAGACCGAGAGAAUUCUCGGUA-CCGUUCUCGUUGUACUCUUCC-CUUUUCGC-CCCGUUUUCGGCACGUCUCGUCGCAUUUUAGU .....(((((.....((.((((((((((((((((((......)))))..-....))))))))))))).)).-......((-(........))).......)))))........ ( -42.10) >DroEre_CAF1 13675 113 - 1 GUCAAGCGGCGCACAGAUGAGUGCAACAAGACCGAGAGAAUUCUCGAUUCCCGUUCUCGUUGUUCUCUUCCCCUUUUCGUCCCCGUUUUCGGCACGUCUCGUCGCAUUUUAGU .....((((((...(((((((.(((((.(((.((.(.(((((...)))))))).))).))))).)))...........((..(((....))).))))))))))))........ ( -30.20) >DroYak_CAF1 13945 112 - 1 GUCAAGCGGCGCACAGAAGAGUACAGCGAGACCGAGAGAAUUCUCGGUUCCCGUUCUCGUUGUUCUGUUCC-CUUUUCGCCCCCGUUUUCGGCACGUCUCGUCGCAUUUUAGU .....((((((.((.(((.((.((((((((((((((......))))).......))))))))).)).))).-......(((.........)))..))..))))))........ ( -38.11) >consensus GUCAAGCGGCACACAGAUGAGUGCAACGAGACCGAGAGAAUUCUCGGUA_CCGUUCUCGUUGUACUCUUCC_CUUUUCGC_CCCGUUUUCGGCACGUCUCGUCGCAUUUUAGU .....(((((.....((.(((.((((((((((((((......))))).......))))))))).))).)).......((...(((....)))..))....)))))........ (-32.61 = -33.09 + 0.48)


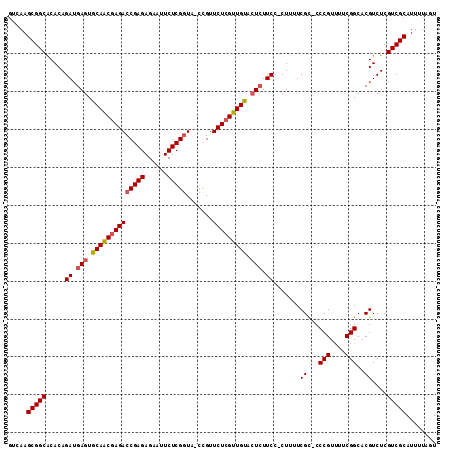
| Location | 9,529,571 – 9,529,670 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -22.67 |
| Energy contribution | -22.95 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
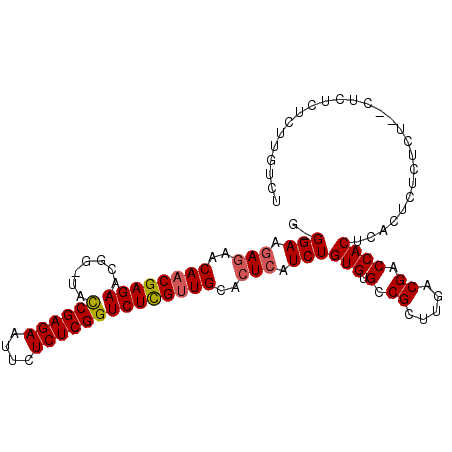
>X_DroMel_CAF1 9529571 99 + 22224390 G-GGAAGA--ACAACGAGAACAG-UACCGAGAAUUCUCUCGGUCUCGUUGCACUCAUCUGUGUGCCGCUUGACGACCACUCACUCUCUCU--CUCUCUCUUGUCU (-(.((((--.....((((..((-((((((((....))))))).(((((((((.(....).)))).....)))))......)))...)))--)....)))).)). ( -25.90) >DroSec_CAF1 13406 103 + 1 G-GGAAGAGUACAACGAGAACGG-UACCGAGAAUUCUCUCGGUCUCGUUGCACUCAUCUGUGUGCCGCUUGACGACCACUCACUCUCUCUCUCUCUCUCUUGUCU (-((((((((....((((((((.-.(((((((....)))))))..))))((((.(....).))))..))))..((....))))))).)))).............. ( -31.80) >DroSim_CAF1 12909 103 + 1 G-GGAAGAGUACAACGAGAACGG-UACCGAGAAUUCUCUCGGUCUCGUUGCACUCAUCUGUGUGCCGCUUGACGACCACUCACUCUCUCUCUCUCUCUCUUGUCU (-((((((((....((((((((.-.(((((((....)))))))..))))((((.(....).))))..))))..((....))))))).)))).............. ( -31.80) >DroEre_CAF1 13715 99 + 1 GGGGAAGAGAACAACGAGAACGGGAAUCGAGAAUUCUCUCGGUCUUGUUGCACUCAUCUGUGCGCCGCUUGACGACCACUCGCUCUC------UCUCUCUUCUCU ((((((((((.....((((.((((...(((((....)))))(((..(((((((......)))))..))..))).....)))).))))------..)))))))))) ( -38.60) >DroYak_CAF1 13985 98 + 1 G-GGAACAGAACAACGAGAACGGGAACCGAGAAUUCUCUCGGUCUCGCUGUACUCUUCUGUGCGCCGCUUGACGACCACUCGCUCUC------UCUCUCUUGUAU .-((.((((((....(((.(((((((((((((....))))))).)).)))).)))))))))...))....(((((....))).))..------............ ( -28.60) >consensus G_GGAAGAGAACAACGAGAACGG_UACCGAGAAUUCUCUCGGUCUCGUUGCACUCAUCUGUGUGCCGCUUGACGACCACUCACUCUCUCU__CUCUCUCUUGUCU ..(((.(((..((((((((.......((((((....))))))))))))))..))).)))(((.(.((.....)).)))).......................... (-22.67 = -22.95 + 0.28)


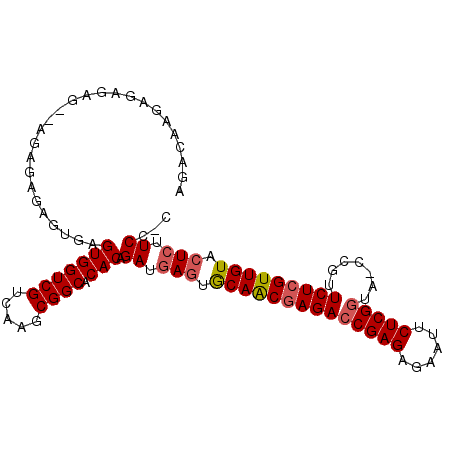
| Location | 9,529,571 – 9,529,670 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -29.81 |
| Energy contribution | -30.29 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9529571 99 - 22224390 AGACAAGAGAGAG--AGAGAGAGUGAGUGGUCGUCAAGCGGCACACAGAUGAGUGCAACGAGACCGAGAGAAUUCUCGGUA-CUGUUCUCGUUGU--UCUUCC-C .............--.(..(((....(((((((.....)))).)))........((((((((((((((......)))))..-....)))))))))--)))..)-. ( -29.90) >DroSec_CAF1 13406 103 - 1 AGACAAGAGAGAGAGAGAGAGAGUGAGUGGUCGUCAAGCGGCACACAGAUGAGUGCAACGAGACCGAGAGAAUUCUCGGUA-CCGUUCUCGUUGUACUCUUCC-C ..........................(((((((.....)))).))).((.((((((((((((((((((......)))))..-....))))))))))))).)).-. ( -37.70) >DroSim_CAF1 12909 103 - 1 AGACAAGAGAGAGAGAGAGAGAGUGAGUGGUCGUCAAGCGGCACACAGAUGAGUGCAACGAGACCGAGAGAAUUCUCGGUA-CCGUUCUCGUUGUACUCUUCC-C ..........................(((((((.....)))).))).((.((((((((((((((((((......)))))..-....))))))))))))).)).-. ( -37.70) >DroEre_CAF1 13715 99 - 1 AGAGAAGAGAGA------GAGAGCGAGUGGUCGUCAAGCGGCGCACAGAUGAGUGCAACAAGACCGAGAGAAUUCUCGAUUCCCGUUCUCGUUGUUCUCUUCCCC ...((((((((.------(((((((...(((((((....)))((((......)))).....))))(((......)))......)))))))....))))))))... ( -39.10) >DroYak_CAF1 13985 98 - 1 AUACAAGAGAGA------GAGAGCGAGUGGUCGUCAAGCGGCGCACAGAAGAGUACAGCGAGACCGAGAGAAUUCUCGGUUCCCGUUCUCGUUGUUCUGUUCC-C ....((.((((.------(((((((.(((((((.....)))).))).............(..((((((......))))))..))))))))....)))).))..-. ( -33.60) >consensus AGACAAGAGAGAG__AGAGAGAGUGAGUGGUCGUCAAGCGGCACACAGAUGAGUGCAACGAGACCGAGAGAAUUCUCGGUA_CCGUUCUCGUUGUACUCUUCC_C ..........................(((((((.....)))).))).((.(((.((((((((((((((......))))).......))))))))).))).))... (-29.81 = -30.29 + 0.48)

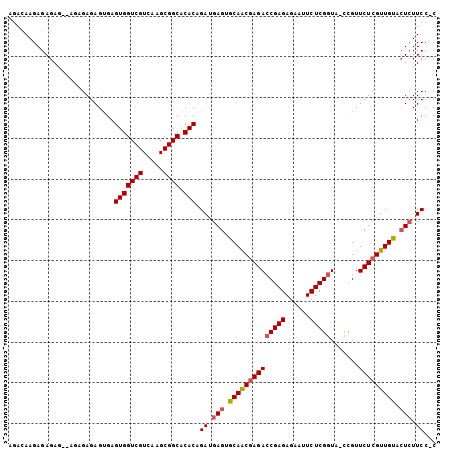
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:20 2006