
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,527,263 – 9,527,404 |
| Length | 141 |
| Max. P | 0.922446 |

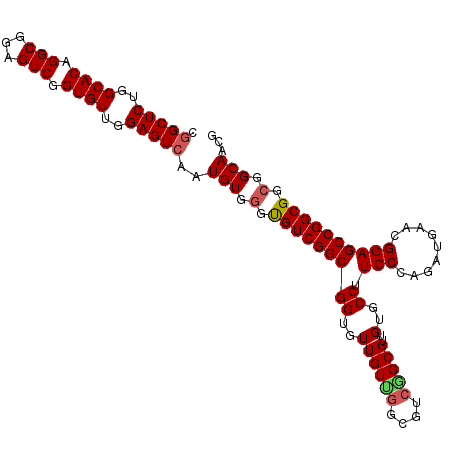
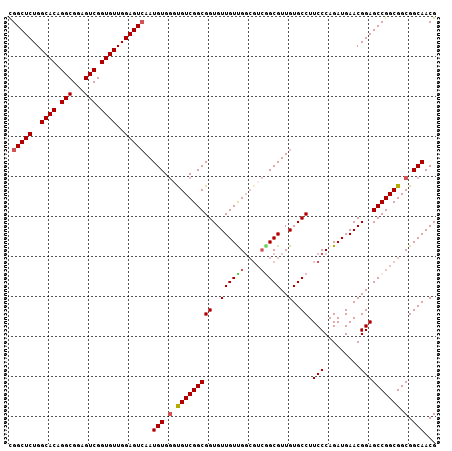
| Location | 9,527,263 – 9,527,370 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -49.90 |
| Consensus MFE | -42.22 |
| Energy contribution | -42.54 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9527263 107 - 22224390 CGGCUCUGGCACAGGCGGAGUCGGUGUUGGAGUGAAUGUGGGUGUCGGCGGUGUUGUUGGCGUCGGCGUUGUGCCUUCCCAGAUGAACGGAGCCGGCGGCGGCAACG ..((((..((((.(((...))).))))..))))...(((.(.(((((((..((((.((((.(..(((.....)))..)))))...))))..))))))).).)))... ( -46.00) >DroSec_CAF1 11013 107 - 1 CGGCUCUGGCACAGGCGGAGUCGGUGUUGGAGUCAAUGUGGGUGUCGGCGGUGUUGUUGGCGUCAGCGUUGUGCCUUCCCAGAUGAACGGAGCCGGCGGCGGCAACG .(((((..((((.(((...))).))))..)))))..(((.(.(((((((((..((((((....)))))..)..)).(((.........)))))))))).).)))... ( -45.10) >DroSim_CAF1 10501 107 - 1 CGGCUCUGGCACAGGCGGAGUCGGUGUUGGAGUCAAUGUGGGUGUCGGCGGUGUUGUUGGCGUCGGCGUUGUGCCUUCCCAGAUGAACGGAGCCGGCGGCGGCAACG .(((((..((((.(((...))).))))..)))))..(((.(.(((((((..((((.((((.(..(((.....)))..)))))...))))..))))))).).)))... ( -48.90) >DroEre_CAF1 11137 101 - 1 CGGCUCUGGCACUGGCGGAGUCGGUGUUGGAGUCAAUGUGACCGUCGGCGGUGUUGUCG------GCGUUGUGCCUUCCCAGAUGAACGGAGCCGGCGGCGGCAACG .(((((..((((((((...))))))))..)))))......((((....))))(((((((------.(((((.((..(((.........)))))))))).))))))). ( -49.70) >DroYak_CAF1 11318 107 - 1 UGGCUCUGGCACUGGCGGAGUCGGUGUUGGAGUCAAUGUGGCCGUCGGCGGUGUUGUCGGCGUCGGCGUUGUGCCUUCCCAGAUGAACGGAGCCGGCGGCGGCAACG ((((((..((((((((...))))))))..)))))).(((.(((((((((..(((((((((.(..(((.....)))..))).))).))))..))))))))).)))... ( -59.80) >consensus CGGCUCUGGCACAGGCGGAGUCGGUGUUGGAGUCAAUGUGGGUGUCGGCGGUGUUGUUGGCGUCGGCGUUGUGCCUUCCCAGAUGAACGGAGCCGGCGGCGGCAACG .(((((..((((.(((...))).))))..)))))..(((.(.(((((((((..((((((....)))))..)..)).(((.........)))))))))).).)))... (-42.22 = -42.54 + 0.32)

| Location | 9,527,302 – 9,527,404 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.05 |
| Mean single sequence MFE | -50.35 |
| Consensus MFE | -30.11 |
| Energy contribution | -30.83 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9527302 102 - 22224390 GUGGCGGACCCAACAAGCCGGCAGCCCAAGGACC------CGGCUCUGGCACAGGCGGAGUCGGUGUUGGAGUG------------AAUGUGGGUGUCGGCGGUGUUGUUGGCGUCGGCG ...((.(((((((((((((((((.((((..((((------((((((((.(...).))))))))).)))......------------....)))))))))))....))))))).))).)). ( -51.40) >DroSec_CAF1 11052 102 - 1 GUGGAGGACCCAACAAGCCGGCAGCCCAAGGACC------CGGCUCUGGCACAGGCGGAGUCGGUGUUGGAGUC------------AAUGUGGGUGUCGGCGGUGUUGUUGGCGUCAGCG ...(..(((((((((((((((((.((((.(....------)(((((..((((.(((...))).))))..)))))------------....)))))))))))....))))))).)))..). ( -50.00) >DroSim_CAF1 10540 102 - 1 GUGGAGGACCCAACAAGCCGGCAGCCCAAGGACC------CGGCUCUGGCACAGGCGGAGUCGGUGUUGGAGUC------------AAUGUGGGUGUCGGCGGUGUUGUUGGCGUCGGCG ((.((.(.(((((((.(((((((.((((.(....------)(((((..((((.(((...))).))))..)))))------------....)))))))))))..)))))..))).)).)). ( -50.90) >DroEre_CAF1 11176 102 - 1 GUGGUGGACCCAAUAAACCGGCAGCCCAAGGACCCGGUCCCGGCUCUGGCACUGGCGGAGUCGGUGUUGGAGUC------------AAUGUGACCGUCGGCGGUGUUGUCG------GCG .(((.....))).....((((((((((...(((..(((((.(((((..((((((((...))))))))..)))))------------...).)))))))...)).)))))))------).. ( -45.00) >DroYak_CAF1 11357 102 - 1 GUGGCGGACCCAACAAACCGGCAGCCCAAGGACC------UGGCUCUGGCACUGGCGGAGUCGGUGUUGGAGUC------------AAUGUGGCCGUCGGCGGUGUUGUCGGCGUCGGCG ...((.(((((.((((.(((((.(((((......------((((((..((((((((...))))))))..)))))------------).)).))).))))).....)))).)).))).)). ( -48.60) >DroAna_CAF1 16128 114 - 1 GCGGCGGACCAAAUAAGCCACCUGCCCAGGGACCUGGACCCGGUUCUGGACCCGGCGGUGUUGGUGUCGGAGUCGUCGGCGGCGUUAAUGUUGGCGUCG------UCGGCGGCGUCGGCG ..((....))......((((((((((..(((.((.((((...)))).)).)))))))).).))))((((..((((((((((((((((....))))))))------))))))))..)))). ( -56.20) >consensus GUGGCGGACCCAACAAGCCGGCAGCCCAAGGACC______CGGCUCUGGCACAGGCGGAGUCGGUGUUGGAGUC____________AAUGUGGGCGUCGGCGGUGUUGUCGGCGUCGGCG ......((((((((..((((((.(((..((..((.......))..))......)))...))))))))))).)))...............((.(((((((((......))))))))).)). (-30.11 = -30.83 + 0.72)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:15 2006