
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,526,968 – 9,527,125 |
| Length | 157 |
| Max. P | 0.594780 |

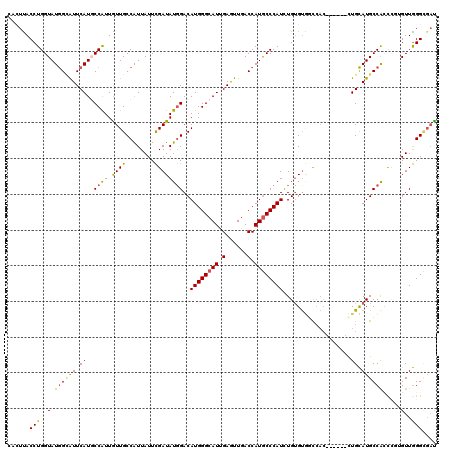
| Location | 9,526,968 – 9,527,088 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -22.42 |
| Energy contribution | -21.82 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9526968 120 + 22224390 UAAUCACCUGGUAUGGCGUUCAUGCCAUUGUUGCCAUUAUUCGAUAUUGACAUGGGCAUUGAGUUGACCAUACCCAUCUGAGUGCCCACCACCUGCUGCAUGCCGCCCUGGUUGGGUGAC ...((((((..(..((((..(((((((.(((((........))))).))...((((((((.((.((........)).)).)))))))).........))))).))))...)..)))))). ( -41.80) >DroVir_CAF1 15981 114 + 1 GAAUUACCUGGUAUAGCAUUCAUGCCAUUAUUGCCAUUAUUUGAUAUGGACAUGGGCAUAGAAUUAACCAUGCCCAUUUGUGUAGUCAC------UUGUAUGCCACCCGUGUUAGGCGAU ........((((((.......))))))..((((((......((((((((..((((((((.(......).))))))))..(((....)))------...........)))))))))))))) ( -29.80) >DroGri_CAF1 11150 114 + 1 GACUUACCUGGUAUGGUAUUCAUGCCAUUGUUGCCAUUAUUCGAUAUGGACAUGGGCAUUGAAUUGACCAUGCCCAUUUGUGUCGCAAC------CUGCAUGCCACCGGUGUUGGGCGAU ........(((((((.....)))))))..((((((......(((((..((..(((((((.(......).)))))))))..)))))((((------(((........))).)))))))))) ( -37.90) >DroWil_CAF1 11369 114 + 1 CUCUUACCUGGUAUGGCGUUCAUUCCAUUAUUGCCAUUAUUCGAUAUGGACAUGGGCAUUGAGUUGACCAUGCCCAUUUGUGCGGCC---ACUUGCUGCAUGCCAGU---GUUGGGUGAU ...((((((..((((((......(((((.((((........))))))))).((((((((.(......).))))))))..(((((((.---....))))))))))).)---)..)))))). ( -40.70) >DroMoj_CAF1 11729 112 + 1 --CUUACCUGGUAUGGAAUUCAUGCCGUUGUUGCCAUUAUUCGAUAUGGACAUGGGCAUUGAGUUGACCAUGCCCAUCUGUGUGGCCAC------CUGCAUGCCAGCCGUGUUGGGCGAU --.(..(((..(((((....(((((....((.(((((....((..((((.(((((.((......)).))))).)))).)).))))).))------..)))))....)))))..)))..). ( -37.80) >DroAna_CAF1 15749 120 + 1 CACUCACCUGGUAUGGCAUUCAUUCCGUUGUUGCCGUUAUUCGAUAUGGACAUGGGCAUCGAGUUGACCAUGCCCAUCUGUGUGCCCACCACCUGCUGCAUGCCGCCCUGGUUGGGACUC ..((((.(..(..((((((.((.(((((.((((........)))))))))..(((((((..((.((........)).))..)))))))........)).))))))..)..).)))).... ( -35.20) >consensus CACUUACCUGGUAUGGCAUUCAUGCCAUUGUUGCCAUUAUUCGAUAUGGACAUGGGCAUUGAGUUGACCAUGCCCAUCUGUGUGGCCAC______CUGCAUGCCACCCGUGUUGGGCGAU ......(((..(.((((((.((..((((.((((........))))))))..((((((((.(......).))))))))...................)).)))))).....)..))).... (-22.42 = -21.82 + -0.61)


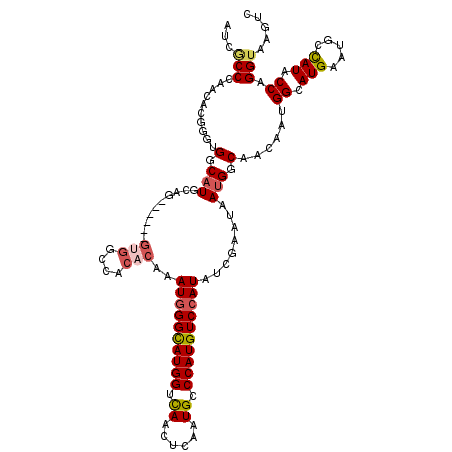
| Location | 9,526,968 – 9,527,088 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -20.82 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9526968 120 - 22224390 GUCACCCAACCAGGGCGGCAUGCAGCAGGUGGUGGGCACUCAGAUGGGUAUGGUCAACUCAAUGCCCAUGUCAAUAUCGAAUAAUGGCAACAAUGGCAUGAACGCCAUACCAGGUGAUUA ((((((.......((((.(((((....((((...((((......(((((((.(......).)))))))))))..))))......((....))...)))))..))))......)))))).. ( -38.02) >DroVir_CAF1 15981 114 - 1 AUCGCCUAACACGGGUGGCAUACAA------GUGACUACACAAAUGGGCAUGGUUAAUUCUAUGCCCAUGUCCAUAUCAAAUAAUGGCAAUAAUGGCAUGAAUGCUAUACCAGGUAAUUC ...((((.....(((((((((....------(((.(((.....((((((((((......))))))))))(.((((........))))).....))))))..))))))).))))))..... ( -33.40) >DroGri_CAF1 11150 114 - 1 AUCGCCCAACACCGGUGGCAUGCAG------GUUGCGACACAAAUGGGCAUGGUCAAUUCAAUGCCCAUGUCCAUAUCGAAUAAUGGCAACAAUGGCAUGAAUACCAUACCAGGUAAGUC .(((((.......)))))(((((..------(((((((((....(((((((((.....)).)))))))))))(((........))))))))....)))))..((((......)))).... ( -35.90) >DroWil_CAF1 11369 114 - 1 AUCACCCAAC---ACUGGCAUGCAGCAAGU---GGCCGCACAAAUGGGCAUGGUCAACUCAAUGCCCAUGUCCAUAUCGAAUAAUGGCAAUAAUGGAAUGAACGCCAUACCAGGUAAGAG ...(((...(---(((.((.....)).)))---)((((.....((((((((((.((......)).)))))))))).........)))).....(((.(((.....))).))))))..... ( -31.64) >DroMoj_CAF1 11729 112 - 1 AUCGCCCAACACGGCUGGCAUGCAG------GUGGCCACACAGAUGGGCAUGGUCAACUCAAUGCCCAUGUCCAUAUCGAAUAAUGGCAACAACGGCAUGAAUUCCAUACCAGGUAAG-- ((((((......)))((((((((..------((.((((.....((((((((((.((......)).)))))))))).........)))).))....)))))....))).....)))...-- ( -34.44) >DroAna_CAF1 15749 120 - 1 GAGUCCCAACCAGGGCGGCAUGCAGCAGGUGGUGGGCACACAGAUGGGCAUGGUCAACUCGAUGCCCAUGUCCAUAUCGAAUAACGGCAACAACGGAAUGAAUGCCAUACCAGGUGAGUG ..((((......))))..(((.((.(.((((...((((..((.((((((((((.((......)).))))))))))..........(....).......))..)))).)))).).)).))) ( -37.30) >consensus AUCGCCCAACACGGGUGGCAUGCAG______GUGGCCACACAAAUGGGCAUGGUCAACUCAAUGCCCAUGUCCAUAUCGAAUAAUGGCAACAAUGGCAUGAAUGCCAUACCAGGUAAGUC ...(((..........(.(((..........(((....)))..((((((((((.((......)).))))))))))........))).)......((.(((.....))).)).)))..... (-20.82 = -21.40 + 0.58)

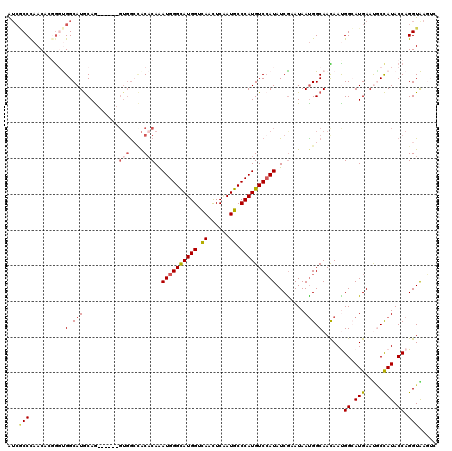
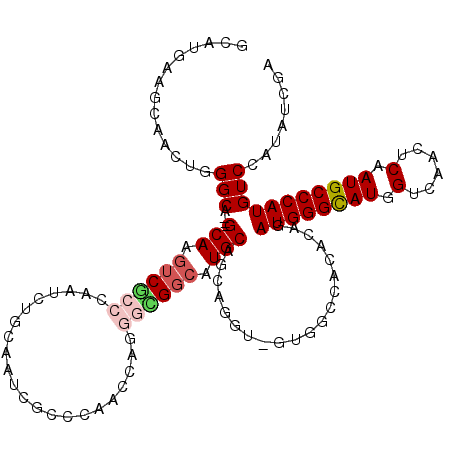
| Location | 9,527,008 – 9,527,125 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -22.16 |
| Energy contribution | -22.85 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9527008 117 - 22224390 GCAUGCAGCAGCUGGGCA---GCAAGUCGCCCAACCUGCAGUCACCCAACCAGGGCGGCAUGCAGCAGGUGGUGGGCACUCAGAUGGGUAUGGUCAACUCAAUGCCCAUGUCAAUAUCGA ((.((((((((.(((((.---.......)))))..)))).(((.(((.....))).))).)))))).((((...((((......(((((((.(......).)))))))))))..)))).. ( -44.30) >DroGri_CAF1 11190 111 - 1 GCAUGAAUCAACUUGGCA---GCAAGUCACCCAAUUUACAAUCGCCCAACACCGGUGGCAUGCAG------GUUGCGACACAAAUGGGCAUGGUCAAUUCAAUGCCCAUGUCCAUAUCGA ...(((..(((((((.((---....((((((......................)))))).)))))------))))........((((((((((.((......)).)))))))))).))). ( -31.05) >DroWil_CAF1 11409 114 - 1 GAAUGCAACAAUUGGGCAGUAGCAAGUCACCGAAUCUACAAUCACCCAAC---ACUGGCAUGCAGCAAGU---GGCCGCACAAAUGGGCAUGGUCAACUCAAUGCCCAUGUCCAUAUCGA ((.(((.....(((((..((((....((...))..)))).....)))))(---(((.((.....)).)))---)...)))...((((((((((.((......)).)))))))))).)).. ( -31.80) >DroMoj_CAF1 11767 111 - 1 GCAUGAAUCAACUGGGCA---GCAAGUCGCCUAACCUGCAAUCGCCCAACACGGCUGGCAUGCAG------GUGGCCACACAGAUGGGCAUGGUCAACUCAAUGCCCAUGUCCAUAUCGA ...(((.....((((((.---....)))(((..(((((((.(((((......))).))..)))))------)))))....)))((((((((((.((......)).)))))))))).))). ( -39.70) >DroAna_CAF1 15789 117 - 1 GGAUGCAGCAACUGGGCA---GCAAGUCGCCCAAUCUGCAGAGUCCCAACCAGGGCGGCAUGCAGCAGGUGGUGGGCACACAGAUGGGCAUGGUCAACUCGAUGCCCAUGUCCAUAUCGA ((((((((....(((((.---.......)))))..))))...((((((.(((..((........))...))))))).))....((((((((.(......).))))))))))))....... ( -45.20) >DroPer_CAF1 8765 114 - 1 GGAUGAACCAAUUGGGCA---GCAAAUCCCCAAAUCUGCAGUCGCCCAAUCAGAGCGUCAUGCAGCAGGU---UGCCACACAGAUGGGCAUGGUAAAUUCGAUGCCCAUGUCGAUAUCGA ((...((((..(((((.(---.....).)))))..((((((.(((.(.....).))).).)))))..)))---).))......((((((((.(......).)))))))).(((....))) ( -36.70) >consensus GCAUGAAGCAACUGGGCA___GCAAGUCGCCCAAUCUGCAAUCGCCCAACCAGGGCGGCAUGCAGCAGGU_GUGGCCACACAGAUGGGCAUGGUCAACUCAAUGCCCAUGUCCAUAUCGA ..............(((....(((.((((((......................)))))).)))....................((((((((.(......).)))))))))))........ (-22.16 = -22.85 + 0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:13 2006