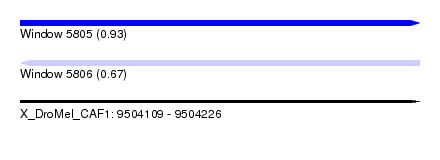
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,504,109 – 9,504,226 |
| Length | 117 |
| Max. P | 0.929296 |

| Location | 9,504,109 – 9,504,226 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.35 |
| Mean single sequence MFE | -40.26 |
| Consensus MFE | -26.18 |
| Energy contribution | -26.14 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
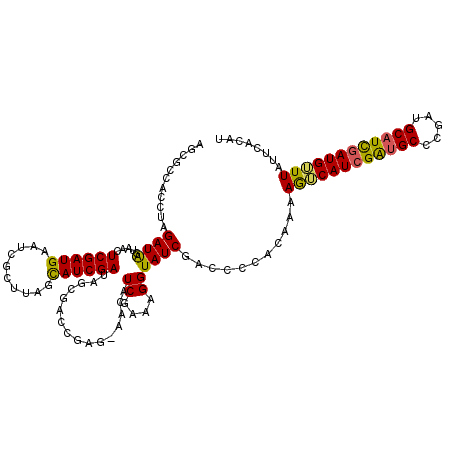
>X_DroMel_CAF1 9504109 117 + 22224390 AUGUGAAUAAACAUCGAUGCAUUGGGCAUCGAUGACUUUUGUGGGGUCGAUACCUUUCGAUGUU-CUCGGUCGUUAUCGAUGCUAAGCGAUGCAUCGAUUUACUAUCUAGGUGGAGCU .(((......)))(((((((((((((((((((((((..(((.((((((((......))))).))-).)))..))))))))))))...)))))))))))((((((.....))))))... ( -45.40) >DroSec_CAF1 47436 117 + 1 AUGUGAAUAACCAUCGAAGCAUCGGGCACCGAUGACUUUUGUGAGGUCGAUACCUUUCGAAGUU-CUCGGUCGCUAUCGAUGCUAAGCGAUUCAUCGAGUUACUAUCUAGGUGGCGCU ..(((.....(((((..(((((((((((((((.((((((...(((((....)))))..))))))-.))))).)))..))))))).((..((((...))))..)).....)))))))). ( -40.10) >DroSim_CAF1 48480 117 + 1 AUGUGAAUAACCAUCGAUGCAUCGGGCACCGAUGACUUUUGUGGGGUCGAUACCUUUCGAAGUU-CUCGGUCGCUAUCGAUGCUAAGCGAUUCAUCGAGUUACUAUCUAGGUGGCGCU ..((((......((((..((((((((((((((.((((((.(..((((....))))..)))))))-.))))).)))..))))))....))))))))...((((((.....))))))... ( -37.90) >DroEre_CAF1 47495 118 + 1 AUGUGAAUAAACAUCAAUGCAUCGGGCAUCGAUGUUUUUUCGGCUCUCGAUACCUUUCGAUGUUUGCCCUUGUCUAUCGAUAUUAAGCGAUUCAUCGAGUUACCAUCUAGGUGGCGCA ...((((.(((((((.((((.....)))).))))))).))))((............((((((.((((...((((....))))....))))..))))))((((((.....)))))))). ( -36.80) >DroYak_CAF1 48103 116 + 1 AUGUGAAUAAACAUGGAUGCAUCGGGCAUCGAUGUUU-UUCGGGUAUCGAUACCUUUCGAUACA-CCGCUUCGCUAUCGAUGUUCAGCGAUUCAUCGAGUUACUAUCUAGGUGGCGCA ..(((((.((((((.(((((.....))))).))))))-..((((((((((......))))))).-))).)))))..((((((..........))))))((((((.....))))))... ( -41.10) >consensus AUGUGAAUAAACAUCGAUGCAUCGGGCAUCGAUGACUUUUGUGGGGUCGAUACCUUUCGAUGUU_CUCGGUCGCUAUCGAUGCUAAGCGAUUCAUCGAGUUACUAUCUAGGUGGCGCU .............((((((.((((((((((((((.......((((.((((......)))).....)))).....))))))))))...)))).))))))((((((.....))))))... (-26.18 = -26.14 + -0.04)


| Location | 9,504,109 – 9,504,226 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.35 |
| Mean single sequence MFE | -32.06 |
| Consensus MFE | -22.14 |
| Energy contribution | -21.46 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9504109 117 - 22224390 AGCUCCACCUAGAUAGUAAAUCGAUGCAUCGCUUAGCAUCGAUAACGACCGAG-AACAUCGAAAGGUAUCGACCCCACAAAAGUCAUCGAUGCCCAAUGCAUCGAUGUUUAUUCACAU ...........(((((...((((((((((.(....((((((((.((...(((.-....)))...(((....)))........)).)))))))).).))))))))))..)))))..... ( -31.90) >DroSec_CAF1 47436 117 - 1 AGCGCCACCUAGAUAGUAACUCGAUGAAUCGCUUAGCAUCGAUAGCGACCGAG-AACUUCGAAAGGUAUCGACCUCACAAAAGUCAUCGGUGCCCGAUGCUUCGAUGGUUAUUCACAU ...........((..((((((......((((...(((((((...((.(((((.-.((((....((((....)))).....))))..))))))).))))))).)))))))))))).... ( -31.40) >DroSim_CAF1 48480 117 - 1 AGCGCCACCUAGAUAGUAACUCGAUGAAUCGCUUAGCAUCGAUAGCGACCGAG-AACUUCGAAAGGUAUCGACCCCACAAAAGUCAUCGGUGCCCGAUGCAUCGAUGGUUAUUCACAU ...........((..(((((((((((.((((....((((((((..(...((((-...))))...(((....)))........)..)))))))).)))).))))))..))))))).... ( -29.60) >DroEre_CAF1 47495 118 - 1 UGCGCCACCUAGAUGGUAACUCGAUGAAUCGCUUAAUAUCGAUAGACAAGGGCAAACAUCGAAAGGUAUCGAGAGCCGAAAAAACAUCGAUGCCCGAUGCAUUGAUGUUUAUUCACAU .((((((......))))..((((((.....((((....((....))...))))....(((....))))))))).)).(((.((((((((((((.....)))))))))))).))).... ( -36.00) >DroYak_CAF1 48103 116 - 1 UGCGCCACCUAGAUAGUAACUCGAUGAAUCGCUGAACAUCGAUAGCGAAGCGG-UGUAUCGAAAGGUAUCGAUACCCGAA-AAACAUCGAUGCCCGAUGCAUCCAUGUUUAUUCACAU .((........((.......))......((((((........)))))).))((-.(((((((......)))))))))(((-((((((.(((((.....))))).)))))).))).... ( -31.40) >consensus AGCGCCACCUAGAUAGUAACUCGAUGAAUCGCUUAGCAUCGAUAGCGACCGAG_AACAUCGAAAGGUAUCGACCCCACAAAAGUCAUCGAUGCCCGAUGCAUCGAUGUUUAUUCACAU ...........((((.....((((((..........))))))................((....))))))...........((((((((((((.....))))))))))))........ (-22.14 = -21.46 + -0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:06 2006