| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,492,059 – 9,492,150 |
| Length | 91 |
| Max. P | 0.995647 |

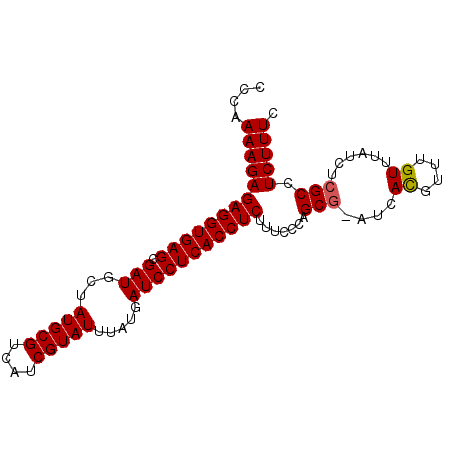
| Location | 9,492,059 – 9,492,150 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 95.62 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -21.98 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9492059 91 + 22224390 CCCAAAAGAGAGGUGAGCGAUGCUAUGCGUCAUCGUAUUUAUGAUCCUCACCUCUUUCCCAGCG-AUCACGUUUGUUUAUCUCGCCUCUUUC ......(((((((((((....((...((((.(((((......((........)).......)))-)).))))..))....))))))))))). ( -26.32) >DroSec_CAF1 35486 91 + 1 CCCAAAAGAGAGGUGAGCGAUGCUAUGCGUCAUCGUAUUUAUGAUCCUCACCUCUUUCACAGCG-AUCACGUUGGUUUAUCUCGCCUCUUUC ......(((((((((((....((((.((((.(((((......((........)).......)))-)).))))))))....))))))))))). ( -27.82) >DroSim_CAF1 35403 92 + 1 CCCAAAAGAGAGGUGAGCGAUGCUAUGCGUCAUCGUAUUUAUGAUCCUCACCUCUUUCACAGCAAAUCACGUUUGUUUAUCUCGCCUCUUUC ......(((((((((((.(((((...)))))(((((....))))).)))))))))))...(((((((...)))))))............... ( -24.70) >DroEre_CAF1 35584 91 + 1 CCCAAAAGAGAGGUGAGCGAUGCUAUGCGUCAUCGUAUUUAUGAUCCUCACCUCUGUUCCAGCG-AUCAUGUUUGUUUAUCUCGCGUCUUUC ....(((((((((((((.(((((...)))))(((((....))))).)))))))).......(((-(..(((......))).)))).))))). ( -26.70) >DroYak_CAF1 35986 91 + 1 CCCAAAAGAGAGGUGAGCGAUGGUAUGCGUCAUCGUAUUUAUGAUCCUCACCUCUUUCCCAGCG-AUCACGUUUGUUUAUCUCGCGUCUUUC ......(((((((((((.(((.(((((((....))))..))).))))))))))))))....(((-(...............))))....... ( -28.06) >consensus CCCAAAAGAGAGGUGAGCGAUGCUAUGCGUCAUCGUAUUUAUGAUCCUCACCUCUUUCCCAGCG_AUCACGUUUGUUUAUCUCGCCUCUUUC ....(((((((((((((.(((...(((((....))))).....))))))))))).......(((....((....))......))).))))). (-21.98 = -22.02 + 0.04)


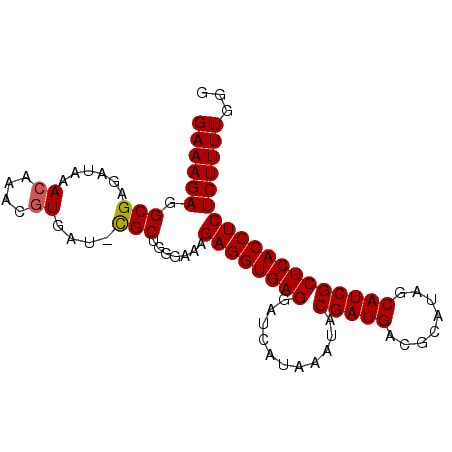
| Location | 9,492,059 – 9,492,150 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 95.62 |
| Mean single sequence MFE | -25.69 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.14 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9492059 91 - 22224390 GAAAGAGGCGAGAUAAACAAACGUGAU-CGCUGGGAAAGAGGUGAGGAUCAUAAAUACGAUGACGCAUAGCAUCGCUCACCUCUCUUUUGGG (((((((((((.....((....))..)-))))......((((((((...........(((((.(.....))))))))))))))))))))... ( -26.80) >DroSec_CAF1 35486 91 - 1 GAAAGAGGCGAGAUAAACCAACGUGAU-CGCUGUGAAAGAGGUGAGGAUCAUAAAUACGAUGACGCAUAGCAUCGCUCACCUCUCUUUUGGG (((((((((((.....((....))..)-))))......((((((((...........(((((.(.....))))))))))))))))))))... ( -26.20) >DroSim_CAF1 35403 92 - 1 GAAAGAGGCGAGAUAAACAAACGUGAUUUGCUGUGAAAGAGGUGAGGAUCAUAAAUACGAUGACGCAUAGCAUCGCUCACCUCUCUUUUGGG ((((((((((((....((....))..))))))......((((((((...........(((((.(.....))))))))))))))))))))... ( -24.90) >DroEre_CAF1 35584 91 - 1 GAAAGACGCGAGAUAAACAAACAUGAU-CGCUGGAACAGAGGUGAGGAUCAUAAAUACGAUGACGCAUAGCAUCGCUCACCUCUCUUUUGGG ((((((.((((...............)-))).......((((((((...........(((((.(.....))))))))))))))))))))... ( -24.36) >DroYak_CAF1 35986 91 - 1 GAAAGACGCGAGAUAAACAAACGUGAU-CGCUGGGAAAGAGGUGAGGAUCAUAAAUACGAUGACGCAUACCAUCGCUCACCUCUCUUUUGGG ....((((((...........)))).)-).(..(((.(((((((((...........(((((........)))))))))))))).)))..). ( -26.20) >consensus GAAAGAGGCGAGAUAAACAAACGUGAU_CGCUGGGAAAGAGGUGAGGAUCAUAAAUACGAUGACGCAUAGCAUCGCUCACCUCUCUUUUGGG ((((((.(((......((....))....))).......((((((((...........(((((........)))))))))))))))))))... (-21.10 = -21.14 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:00 2006