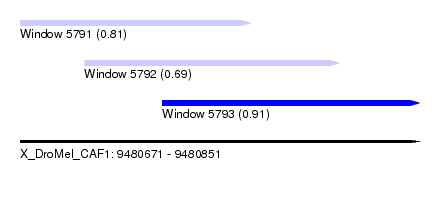
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,480,671 – 9,480,851 |
| Length | 180 |
| Max. P | 0.908370 |

| Location | 9,480,671 – 9,480,775 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -23.48 |
| Consensus MFE | -18.24 |
| Energy contribution | -19.08 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9480671 104 + 22224390 AUUAU--U---------UAAAACACCCAGUUAAGUAACAUUCCUUGAAUUACGCGAUCGGACGUCGAUUU-----UAGCCACGCCUUUUGGAAAUUCCCGAAAUGGCGUAUGGCACAUAU .((((--(---------(((.........)))))))).................(((((.....))))).-----..((((((((((((((......)))))).)))))..)))...... ( -23.00) >DroSec_CAF1 23877 115 + 1 AUUUUAAUCAAAAAUAAAAAAACACCCAAUUAAGUAACAUCCCUUGAAUUACGCGAUCGGACGUCGAUUU-----UAGCCACGCCUUUUGGAAAUUCCCGAAAUGGCGUAUGGCACAUUU ..............(((((..((..((.(((..((((((.....))..))))..))).))..))...)))-----))((((((((((((((......)))))).)))))..)))...... ( -23.10) >DroSim_CAF1 23891 87 + 1 ----------------------------AUUAAGUAACAUCCCUUGAAUUACGCGAUCGGACGUCGAUUU-----UAGCCACGCCUUUUGGAAAUUCCCGAAAUGGCGUAUGGCACAUAU ----------------------------.(((((........))))).......(((((.....))))).-----..((((((((((((((......)))))).)))))..)))...... ( -22.80) >DroEre_CAF1 23953 117 + 1 AGUUCUUACAU---UAAAAAACCACCGAAUUAAGUAAGAUCCCUUUAAUUCGGCGUUCGAAAGUAGAUUUACAUGUAGCCACGCCUUUUGGAAAUACCCGAAAUGACGUAUGGCACAUAU ......(((((---((((....(.((((((((((.........)))))))))).)..(....)....)))).)))))(((((((.((((((......)))))).).)))..)))...... ( -25.00) >DroYak_CAF1 23899 114 + 1 AUUUCUUUCAC-----AAAAACCACCCAAUUAAGUAAAAUCCCUUGAAUUCCGCGAUCGGAAGACGAUUUACACGUGGUCACGCCU-UUGGAAAUUCCCGAAAUGGCGUAUGGCACAUUU ...........-----....(((((....(((((........))))).(((((....)))))............))))).((((((-((((......)))))..)))))........... ( -23.50) >consensus AUUUC__UCA______AAAAAACACCCAAUUAAGUAACAUCCCUUGAAUUACGCGAUCGGACGUCGAUUU_____UAGCCACGCCUUUUGGAAAUUCCCGAAAUGGCGUAUGGCACAUAU .............................(((((........))))).......(((((.....)))))........((((((((((((((......)))))).)))))..)))...... (-18.24 = -19.08 + 0.84)


| Location | 9,480,700 – 9,480,815 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.44 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -24.82 |
| Energy contribution | -25.62 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9480700 115 + 22224390 UCCUUGAAUUACGCGAUCGGACGUCGAUUU-----UAGCCACGCCUUUUGGAAAUUCCCGAAAUGGCGUAUGGCACAUAUUCCAAUGAAUCCAUGCAAUGGCAUUUAAUUGCAAUCUAUA ............(((((((.....))))).-----..((((.(((((((((......)))))).)))((((((..(((......)))...))))))..))))........))........ ( -30.50) >DroSec_CAF1 23917 115 + 1 CCCUUGAAUUACGCGAUCGGACGUCGAUUU-----UAGCCACGCCUUUUGGAAAUUCCCGAAAUGGCGUAUGGCACAUUUUCGAAUGAAUCCGUGCAAUGGCAUUUAAUUGCAAUCUAUA ..............(((((((..((((...-----..((((((((((((((......)))))).)))))..)))......)))).....))))((((((........))))))))).... ( -32.00) >DroSim_CAF1 23903 115 + 1 CCCUUGAAUUACGCGAUCGGACGUCGAUUU-----UAGCCACGCCUUUUGGAAAUUCCCGAAAUGGCGUAUGGCACAUAUUCCAAUGAAUCCAUGCAAUGGCAUUUAAUUGCAAUCUAUA ............(((((((.....))))).-----..((((.(((((((((......)))))).)))((((((..(((......)))...))))))..))))........))........ ( -30.50) >DroEre_CAF1 23990 120 + 1 CCCUUUAAUUCGGCGUUCGAAAGUAGAUUUACAUGUAGCCACGCCUUUUGGAAAUACCCGAAAUGACGUAUGGCACAUAUUCCAUUGAAUCAAUGAGAUGACAUUUAAUUGCAAUCUAUA ........((((.....)))).(((((((....(((.(((((((.((((((......)))))).).)))..))))))(((..(((((...)))))..)))............))))))). ( -24.50) >DroYak_CAF1 23934 119 + 1 CCCUUGAAUUCCGCGAUCGGAAGACGAUUUACACGUGGUCACGCCU-UUGGAAAUUCCCGAAAUGGCGUAUGGCACAUUUUCCAUUGAAUCCAUGAGAUGACAUUUAAUUGCAAUCUAGA ...(((..(((((....)))))..)))......(((((((((((((-((((......)))))..)))))((((........)))).))..)))))((((..((......))..))))... ( -27.80) >consensus CCCUUGAAUUACGCGAUCGGACGUCGAUUU_____UAGCCACGCCUUUUGGAAAUUCCCGAAAUGGCGUAUGGCACAUAUUCCAAUGAAUCCAUGCAAUGGCAUUUAAUUGCAAUCUAUA ............(((((..((.((((...........((((((((((((((......)))))).)))))..))).(((((((....))))..)))...)))).))..)))))........ (-24.82 = -25.62 + 0.80)



| Location | 9,480,735 – 9,480,851 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.65 |
| Mean single sequence MFE | -30.39 |
| Consensus MFE | -23.12 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9480735 116 + 22224390 ACGCCUUUUGGAAAUUCCCGAAAUGGCGUAUGGCACAUAUUCCAAUGAAUCCAUGCAAUGGCAUUUAAUUGCAAUCUAUACAAAUGUCAAGGGGGAACU----CACAAGGAACCCAAUUA (((((((((((......)))))).))))).((((((((......)))......((((((........))))))...........)))))..(((...((----....))...)))..... ( -29.30) >DroSec_CAF1 23952 115 + 1 ACGCCUUUUGGAAAUUCCCGAAAUGGCGUAUGGCACAUUUUCGAAUGAAUCCGUGCAAUGGCAUUUAAUUGCAAUCUAUACAAAUGUCAAGGGGG-ACU----CACAAGGAAGCCAAUUA (((((((((((......)))))).))))).((((.((((....))))..(((.((((((........))))))...........(((...((...-.))----.))).))).)))).... ( -33.20) >DroSim_CAF1 23938 115 + 1 ACGCCUUUUGGAAAUUCCCGAAAUGGCGUAUGGCACAUAUUCCAAUGAAUCCAUGCAAUGGCAUUUAAUUGCAAUCUAUACAAAUGUCAAGGGGG-ACU----CACAAGGAAGCCAAUUA (((((((((((......)))))).))))).((((.....((((..(((.(((.((((((........)))))).(((..((....))..))).))-).)----))...)))))))).... ( -33.30) >DroEre_CAF1 24030 115 + 1 ACGCCUUUUGGAAAUACCCGAAAUGACGUAUGGCACAUAUUCCAUUGAAUCAAUGAGAUGACAUUUAAUUGCAAUCUAUACAAAUGUCAAGGGGG-ACU----CACAAGGAAGCCAAUUA ((((.((((((......)))))).).))).((((.....((((.(((...........((((((((...............)))))))).((...-.))----..))))))))))).... ( -24.46) >DroYak_CAF1 23974 118 + 1 ACGCCU-UUGGAAAUUCCCGAAAUGGCGUAUGGCACAUUUUCCAUUGAAUCCAUGAGAUGACAUUUAAUUGCAAUCUAGACAAAUGUCAAGUGGG-ACUCACUCACAAGGAAGCCAAUUA ((((((-((((......)))))..))))).((((.((........))..(((......((((((((..(((.....)))..)))))))).(((((-.....)))))..))).)))).... ( -31.70) >consensus ACGCCUUUUGGAAAUUCCCGAAAUGGCGUAUGGCACAUAUUCCAAUGAAUCCAUGCAAUGGCAUUUAAUUGCAAUCUAUACAAAUGUCAAGGGGG_ACU____CACAAGGAAGCCAAUUA (((((((((((......)))))).))))).((((.....(((....)))(((.((...((((((((...............))))))))((......))......)).))).)))).... (-23.12 = -23.68 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:54 2006