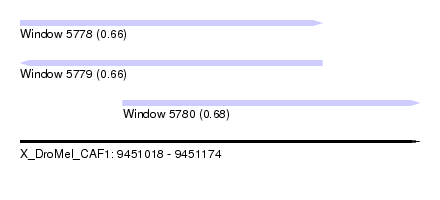
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,451,018 – 9,451,174 |
| Length | 156 |
| Max. P | 0.680519 |

| Location | 9,451,018 – 9,451,136 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.98 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -21.83 |
| Energy contribution | -22.83 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
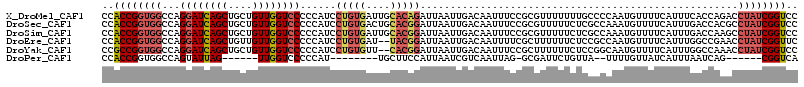
>X_DroMel_CAF1 9451018 118 + 22224390 GCCCGUGUGCGCCUGCCACACCUUAAUGGUGUGGUCAUAACCACCGGUGGCCAGGAUCAGCUGCUGUUGGUCCCCCAUCCUGUGAUUGCACAGAUUAAUUGACAAUUUCCGCGUUUUU .....(((((....((((((((.....))))))))......(((.(((((...((((((((....)))))))).))).)).)))...))))).......................... ( -41.20) >DroPse_CAF1 6853 103 + 1 GCCGGUAUGGGCUUGCCACACCUUGAUUGUGUGAUCGUAUCCACCGGUGGCCAGUAUUAG------UUGGUCCCCCAU--------UGCUUCCAUUAAUCGUCAAUUAG-GCGAUUCU ((((((..(((..((.(((((.......)))))..))..)))))))))((((((......------))))))......--------..........(((((((.....)-)))))).. ( -29.00) >DroSec_CAF1 4643 118 + 1 GCCCGUGUGCGCCUGCCACACCUUUAUGGUGUGGUCAUAACCACCGGUGGCCAGGAUCAGCUGCUGUUGGUCCCCCAUCCUGUGACUGCACGGAUUAAUUGACAAUUUCCGCGUUUUU ...(((((((....((((((((.....))))))))......(((.(((((...((((((((....)))))))).))).)).)))...))))(((.............))))))..... ( -44.02) >DroEre_CAF1 4884 116 + 1 GCCCGUGUGCGCUUGCCACACCUUUAUGGUGUGGUCAUAGCCACCGGUGGCCAGGAUCAGCUGUUGUUGGUCCCCCAUCCUGUGAU--UACGGAUUAAUUGACAAUUUUCGCUUUUUU ..(((((...(((.((((((((.....))))))))...)))(((.(((((...((((((((....)))))))).))).)).)))..--)))))......................... ( -43.20) >DroYak_CAF1 4634 116 + 1 GCCCGUGUGCGCCUGCCACACCUUUAUUGUGUGGUCAUAGCCGCCGGUGGCCAGGAUCAGCUGCUGUUGGUCCCCCAUCCUGUGUU--CACGGAUUAAUUGACAAUUUCCGCUUUUUU ..(((((.((((..(((((((.......)))))))..........(((((...((((((((....)))))))).)))))..)))).--)))))......................... ( -38.90) >DroPer_CAF1 4974 103 + 1 GCCGGUAUGGGCUUGCCAAACCUUGAUUGUGUGAUCGUAUCCACCGGUGGCCAGUAUUAG------UUGGUCCCCCAU--------UGCUUCCAUUAAUCGUCAAUUAG-GCGAUUCU ...((.(((((...(((((......((((.((.((((.......)))).)))))).....------)))))..)))))--------.....))...(((((((.....)-)))))).. ( -28.70) >consensus GCCCGUGUGCGCCUGCCACACCUUUAUGGUGUGGUCAUAACCACCGGUGGCCAGGAUCAGCUGCUGUUGGUCCCCCAUCCUGUGAUUGCACGGAUUAAUUGACAAUUUCCGCGUUUUU (((.(((.((....(((((((.......)))))))....))))).)))((...((((((((....)))))))).)).......................................... (-21.83 = -22.83 + 1.00)

| Location | 9,451,018 – 9,451,136 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.98 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -21.96 |
| Energy contribution | -22.85 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9451018 118 - 22224390 AAAAACGCGGAAAUUGUCAAUUAAUCUGUGCAAUCACAGGAUGGGGGACCAACAGCAGCUGAUCCUGGCCACCGGUGGUUAUGACCACACCAUUAAGGUGUGGCAGGCGCACACGGGC .....((((((..(((.....)))))))))......((((((((....))..(((...)))))))))....(((((((((.((.(((((((.....)))))))))))).))).))).. ( -41.40) >DroPse_CAF1 6853 103 - 1 AGAAUCGC-CUAAUUGACGAUUAAUGGAAGCA--------AUGGGGGACCAA------CUAAUACUGGCCACCGGUGGAUACGAUCACACAAUCAAGGUGUGGCAAGCCCAUACCGGC ......((-(........((((....((....--------.(((....))).------....((((((...)))))).......))....))))..(((((((.....)))))))))) ( -27.80) >DroSec_CAF1 4643 118 - 1 AAAAACGCGGAAAUUGUCAAUUAAUCCGUGCAGUCACAGGAUGGGGGACCAACAGCAGCUGAUCCUGGCCACCGGUGGUUAUGACCACACCAUAAAGGUGUGGCAGGCGCACACGGGC .....((((((..(((.....)))))))))......((((((((....))..(((...)))))))))....(((((((((.((.(((((((.....)))))))))))).))).))).. ( -43.60) >DroEre_CAF1 4884 116 - 1 AAAAAAGCGAAAAUUGUCAAUUAAUCCGUA--AUCACAGGAUGGGGGACCAACAACAGCUGAUCCUGGCCACCGGUGGCUAUGACCACACCAUAAAGGUGUGGCAAGCGCACACGGGC ......((((......))............--....((((((((....))..((.....))))))))))..(((((((((.((.(((((((.....)))))))))))).))).))).. ( -39.20) >DroYak_CAF1 4634 116 - 1 AAAAAAGCGGAAAUUGUCAAUUAAUCCGUG--AACACAGGAUGGGGGACCAACAGCAGCUGAUCCUGGCCACCGGCGGCUAUGACCACACAAUAAAGGUGUGGCAGGCGCACACGGGC ......(((((..(((.....)))))))).--....((((((((....))..(((...)))))))))(((....(((.((..(.((((((.......))))))))).))).....))) ( -38.10) >DroPer_CAF1 4974 103 - 1 AGAAUCGC-CUAAUUGACGAUUAAUGGAAGCA--------AUGGGGGACCAA------CUAAUACUGGCCACCGGUGGAUACGAUCACACAAUCAAGGUUUGGCAAGCCCAUACCGGC ......((-(........((((....((....--------.(((....))).------....((((((...)))))).......))....))))..(((.(((.....))).)))))) ( -23.90) >consensus AAAAACGCGGAAAUUGUCAAUUAAUCCGUGCAAUCACAGGAUGGGGGACCAACAGCAGCUGAUCCUGGCCACCGGUGGAUAUGACCACACAAUAAAGGUGUGGCAAGCGCACACGGGC ........................((((((...........(((....)))......(((.....((((((....)))))).(.((((((.......))))))).)))...)))))). (-21.96 = -22.85 + 0.89)


| Location | 9,451,058 – 9,451,174 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.98 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -19.37 |
| Energy contribution | -20.58 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
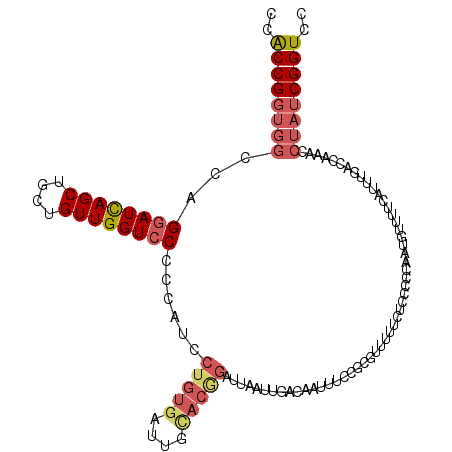
>X_DroMel_CAF1 9451058 116 + 22224390 CCACCGGUGGCCAGGAUCAGCUGCUGUUGGUCCCCCAUCCUGUGAUUGCACAGAUUAAUUGACAAUUUCCGCGUUUUUUUGCCCCAAUGUUUUCAUUUCACCAGACCUAUCGGUCC .....(((((...((((((((....))))))))......(((((....)))))......(((.(((....(((......)))......))).)))..))))).((((....)))). ( -30.20) >DroSec_CAF1 4683 116 + 1 CCACCGGUGGCCAGGAUCAGCUGCUGUUGGUCCCCCAUCCUGUGACUGCACGGAUUAAUUGACAAUUUCCGCGUUUUUCUCGCCAAAUGUUUUCAUUUGACCACGCCUAUCGGUCC ..(((((((((..((((((((....))))))))........(((...((.((((.............)))).)).........((((((....))))))..)))))).)))))).. ( -35.72) >DroSim_CAF1 961 116 + 1 CCACCGGUGGCCAGGAUCAGCUGCUGUUGGUCCCCCAUCCUGUGAUUGCACGGAUUAAUUGACAAUUUCCGCGUUUUUCUCGCCAAAUGUUUUCAUUUGACCAAGCCUAUCGGUCC ..(((((((((..((((((((....))))))))...((((.(((....)))))))...(((.........(((.......)))((((((....))))))..)))))).)))))).. ( -35.50) >DroEre_CAF1 4924 114 + 1 CCACCGGUGGCCAGGAUCAGCUGUUGUUGGUCCCCCAUCCUGUGAU--UACGGAUUAAUUGACAAUUUUCGCUUUUUUCUCCGCCAAUGUUUUCAUUUGGCCGAACCUAUCGGUUC .(((.(((((...((((((((....)))))))).))).)).)))..--...(((..((.(((......)))....))..)))(((((((....)).))))).(((((....))))) ( -32.50) >DroYak_CAF1 4674 114 + 1 CCGCCGGUGGCCAGGAUCAGCUGCUGUUGGUCCCCCAUCCUGUGUU--CACGGAUUAAUUGACAAUUUCCGCUUUUUUCUCCGGCAAUGUUUUCAUUUGGCCAAACCUAUCGGUCC ..(((((((((((((((((((....))))))))...((((.((...--.)))))).....((((.((.(((..........))).))))))......))))))......))))).. ( -30.60) >DroPer_CAF1 5014 93 + 1 CCACCGGUGGCCAGUAUUAG------UUGGUCCCCCAU--------UGCUUCCAUUAAUCGUCAAUUAG-GCGAUUCUGUUA--UUUUGUUAUCAUUUAAUCAG------CGGUCA ..(((((.((((((......------))))))))....--------.(((......(((((((.....)-))))))..((((--...((....))..)))).))------)))).. ( -19.80) >consensus CCACCGGUGGCCAGGAUCAGCUGCUGUUGGUCCCCCAUCCUGUGAUUGCACGGAUUAAUUGACAAUUUCCGCGUUUUUCUCCCCCAAUGUUUUCAUUUGACCAAACCUAUCGGUCC ..((((((((...((((((((....))))))))......(((((....))))).....................................................)))))))).. (-19.37 = -20.58 + 1.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:43 2006