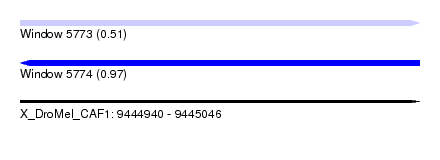
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,444,940 – 9,445,046 |
| Length | 106 |
| Max. P | 0.971913 |

| Location | 9,444,940 – 9,445,046 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 90.09 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -19.19 |
| Energy contribution | -18.87 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9444940 106 + 22224390 AAACCCAUGAGCAUAUGUA--UACAUACUUAUAUGCAUAUGCCCGAGUAUGGUAAUCAAGGCGAACUUAUCAAAGUCAGCCGCACGAAAUUGCUACAAGGCACACGCC ....(((((.(((((((((--((........))))))))))).....))))).......(((..((((....))))..)))(((......))).....(((....))) ( -29.00) >DroEre_CAF1 27230 104 + 1 AUGCCCAUACGCAUAU----GUACAUACUUAUAUGCAUGUGCCCGAGUAUGGUAAUCAAGGCGAACUUAUCAAAGUCAGUCCCACGAAAUUACUACAAGGCACACGCC ..((......((((((----(........))))))).((((((..((((..........(((..((((....))))..))).........))))....)))))).)). ( -24.41) >DroYak_CAF1 26873 108 + 1 UAACCCAUGAGCAUAUGUAUGUACAUACUCAUAUGCAUAUGCCCGAGUAUGGUAAUCAAGGCGAACUUAUCAAAGUCAGCGCCACGAAAUUGCUACAAGGCACACGCC ....(((((.((((((((((((........)))))))))))).....))))).......((((.((((....))))...))))...............(((....))) ( -29.20) >consensus AAACCCAUGAGCAUAUGUA_GUACAUACUUAUAUGCAUAUGCCCGAGUAUGGUAAUCAAGGCGAACUUAUCAAAGUCAGCCCCACGAAAUUGCUACAAGGCACACGCC .....((((.(((((((............))))))).))))........(((((((...(((..((((....))))..))).......)))))))...(((....))) (-19.19 = -18.87 + -0.32)

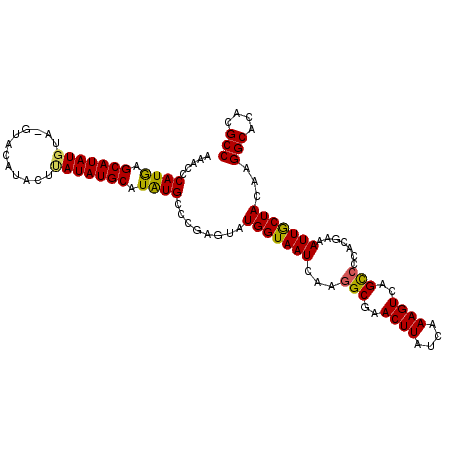

| Location | 9,444,940 – 9,445,046 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 90.09 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -32.24 |
| Energy contribution | -33.13 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9444940 106 - 22224390 GGCGUGUGCCUUGUAGCAAUUUCGUGCGGCUGACUUUGAUAAGUUCGCCUUGAUUACCAUACUCGGGCAUAUGCAUAUAAGUAUGUA--UACAUAUGCUCAUGGGUUU .((((((((((.((((((......)))(((.(((((....))))).)))..........)))..)))))))))).(((.(((((((.--...))))))).)))..... ( -34.10) >DroEre_CAF1 27230 104 - 1 GGCGUGUGCCUUGUAGUAAUUUCGUGGGACUGACUUUGAUAAGUUCGCCUUGAUUACCAUACUCGGGCACAUGCAUAUAAGUAUGUAC----AUAUGCGUAUGGGCAU .((((((((((.(((((((((....(((.(.(((((....))))).)))).))))))..)))..))))))))))........(((..(----(((....))))..))) ( -34.20) >DroYak_CAF1 26873 108 - 1 GGCGUGUGCCUUGUAGCAAUUUCGUGGCGCUGACUUUGAUAAGUUCGCCUUGAUUACCAUACUCGGGCAUAUGCAUAUGAGUAUGUACAUACAUAUGCUCAUGGGUUA .((((((((((.(((......(((.((((..(((((....))))))))).)))......)))..)))))))))).(((((((((((......)))))))))))..... ( -41.30) >consensus GGCGUGUGCCUUGUAGCAAUUUCGUGGGGCUGACUUUGAUAAGUUCGCCUUGAUUACCAUACUCGGGCAUAUGCAUAUAAGUAUGUAC_UACAUAUGCUCAUGGGUUU .((((((((((.(((......(((...(((.(((((....))))).))).)))......)))..)))))))))).(((((((((((......)))))))))))..... (-32.24 = -33.13 + 0.89)

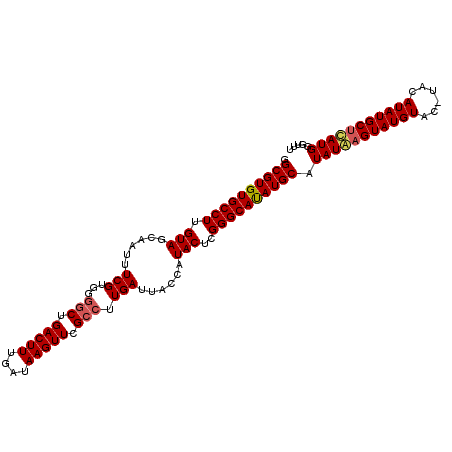

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:37 2006