
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,430,373 – 9,430,549 |
| Length | 176 |
| Max. P | 0.999986 |

| Location | 9,430,373 – 9,430,481 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.53 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -20.55 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
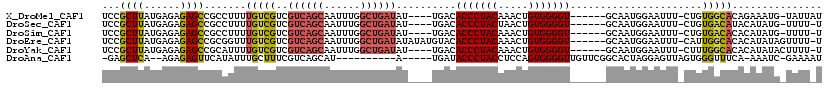
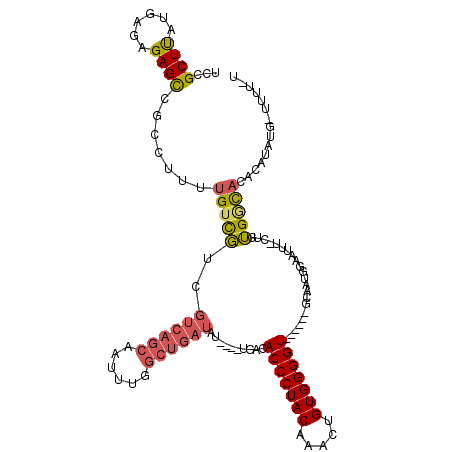
>X_DroMel_CAF1 9430373 108 - 22224390 UCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAAUUUGGCUGAUAU----UGACACCCUACAAACUGUGGGGU------GCAAUGGAAUUU-CUGUGGCACAGAAAUG-UAUUAU (((((((......)))).((.....((((..((((((......))))))..----))))(((((((.....)))))))------))...)))((((-(((.....))))))).-...... ( -38.20) >DroSec_CAF1 12136 107 - 1 UCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAAUUUGGCUGAUAU----UGACACCCUACUAACUGUGGGGU------GCAAUGGAAUUU-CUGUGACAUACAUAUG-UUUU-U (((((((......)))).((.....((((..((((((......))))))..----))))(((((((.....)))))))------))...)))....-....(((((....)))-))..-. ( -33.50) >DroSim_CAF1 12851 107 - 1 UCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAAUUUGGCUGAUAU----UGACACCCUACAAACUGUGGGGU------GCAAUGGAAUUU-CUGUGACACACAUAUG-UUUU-U (((((((......)))).((.....((((..((((((......))))))..----))))(((((((.....)))))))------))...)))....-.((((...))))....-....-. ( -32.80) >DroEre_CAF1 12306 112 - 1 UCCGCUUAUGAGAGAGCCGCGGUUUGUCGUCGUCAGCAAUUUGGCUGAUAUAUAUGUACACCCUACAAACUGUGGGGU------GCAAUGGAAUUU-CAUUGGCACACAUAUAGUUUU-U .((((.............)))).........((((((......)))))).(((((((.((((((((.....)))))))------)((((((....)-)))))....))))))).....-. ( -35.12) >DroYak_CAF1 12795 108 - 1 UCCGCUUAUGAGAGAGCCGCAUUUUGUCGUCGUCAGCAAUUUGGCUGAUAU----UGACACCCUACAAACUGUGGGGU------GCAAUGGAAUUU-CUUUGGCACACAUAUACUUUU-U ...(((...((((((..(.((((..((((..((((((......))))))..----))))(((((((.....)))))))------..)))))..)))-))).)))..............-. ( -35.00) >DroAna_CAF1 9556 100 - 1 -GAGCUCA--AGAGAGUUCAUAUUUGCUUUCGUCAGCAU----------A-----UGAUACCCUACCUCCAGUGGGGUUGUUCGGCACUAGGAGUUAGUGGGUUUCA-AAAUC-GAAAAU -((((((.--...)))))).....((((......)))).----------.-----(((.((((...((((((((..(.....)..)))).)))).....)))).)))-.....-...... ( -27.60) >consensus UCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAAUUUGGCUGAUAU____UGACACCCUACAAACUGUGGGGU______GCAAUGGAAUUU_CUGUGGCACACAUAUG_UUUU_U ...((((......)))).......(((((..((((((......))))))..........(((((((.....)))))))......................)))))............... (-20.55 = -20.88 + 0.34)
| Location | 9,430,407 – 9,430,519 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 95.01 |
| Mean single sequence MFE | -41.12 |
| Consensus MFE | -37.02 |
| Energy contribution | -37.42 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
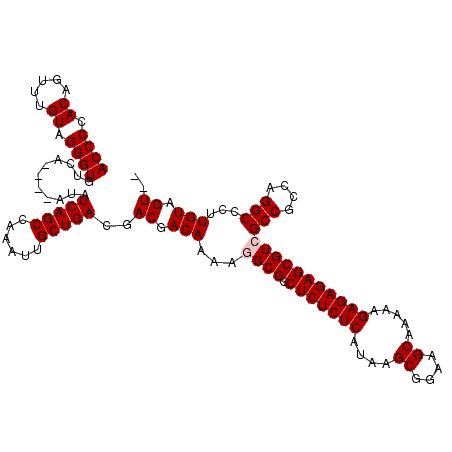
>X_DroMel_CAF1 9430407 112 + 22224390 ACCCCACAGUUUGUAGGGUGUCA----AUAUCAGCCAAAUUGCUGACGACGACAAAAGGCGGCUCUCUCAUAAGCGGAAGCAAAAAGAGAGAGCGUCGCCGCCAGGCCCUUGUAGU-- ((((.((.....)).))))(((.----...(((((......))))).))).((((..((((.(((((((....((....)).....)))))))))))(((....)))..))))...-- ( -41.50) >DroSec_CAF1 12169 111 + 1 ACCCCACAGUUAGUAGGGUGUCA----AUAUCAGCCAAAUUGCUGACGACGACAAAAGGCGGCUCUCUCAUAAGCGGAAGCAAAAAGAGAGAGCGUCGCCGCCAGGCCC-UGUAGU-- .............(((((((((.----...(((((......)))))....)))....((((((((((((....((....)).....))))))))..)))).....))))-))....-- ( -42.40) >DroSim_CAF1 12884 111 + 1 ACCCCACAGUUUGUAGGGUGUCA----AUAUCAGCCAAAUUGCUGACGACGACAAAAGGCGGCUCUCUCAUAAGCGGAAGCAAAAAGAGAGAGCGUCGCCGCCAGGCCC-UGUAGU-- ..........((((((((((((.----...(((((......)))))....)))....((((((((((((....((....)).....))))))))..)))).....))))-))))).-- ( -42.70) >DroEre_CAF1 12340 116 + 1 ACCCCACAGUUUGUAGGGUGUACAUAUAUAUCAGCCAAAUUGCUGACGACGACAAACCGCGGCUCUCUCAUAAGCGGAAGCAAAAAGAGAGAGCGUCGCCGCCAGGCCAUUGUAGU-- ((((.((.....)).)))).((((......(((((......))))).((((......)...((((((((....((....)).....)))))))))))(((....)))...))))..-- ( -38.60) >DroYak_CAF1 12829 114 + 1 ACCCCACAGUUUGUAGGGUGUCA----AUAUCAGCCAAAUUGCUGACGACGACAAAAUGCGGCUCUCUCAUAAGCGGAAGCAAAAAGAGAGAGCGUCGCCGCCAGGCCCUUGUAGUUG ((((.((.....)).))))....----...(((((......)))))((((.((((......((((((((....((....)).....))))))))...(((....)))..)))).)))) ( -40.40) >consensus ACCCCACAGUUUGUAGGGUGUCA____AUAUCAGCCAAAUUGCUGACGACGACAAAAGGCGGCUCUCUCAUAAGCGGAAGCAAAAAGAGAGAGCGUCGCCGCCAGGCCCUUGUAGU__ ((((.((.....)).))))...........(((((......)))))..((.(((...((((.(((((((....((....)).....)))))))))))(((....)))...))).)).. (-37.02 = -37.42 + 0.40)

| Location | 9,430,407 – 9,430,519 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 95.01 |
| Mean single sequence MFE | -46.16 |
| Consensus MFE | -44.08 |
| Energy contribution | -44.68 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.41 |
| SVM RNA-class probability | 0.999986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
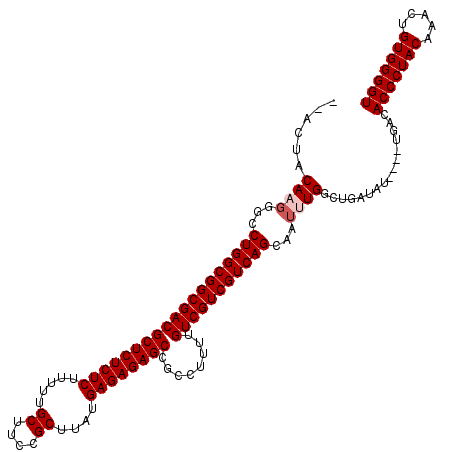
>X_DroMel_CAF1 9430407 112 - 22224390 --ACUACAAGGGCCUGGCGGCGACGCUCUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAAUUUGGCUGAUAU----UGACACCCUACAAACUGUGGGGU --.....(((((((....)))...((((((((.....((....))....))))))))..))))..((((..((((((......))))))..----))))(((((((.....))))))) ( -46.50) >DroSec_CAF1 12169 111 - 1 --ACUACA-GGGCCUGGCGGCGACGCUCUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAAUUUGGCUGAUAU----UGACACCCUACUAACUGUGGGGU --....((-(...)))..((((..((((((((.....((....))....))))))))))))....((((..((((((......))))))..----))))(((((((.....))))))) ( -46.40) >DroSim_CAF1 12884 111 - 1 --ACUACA-GGGCCUGGCGGCGACGCUCUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAAUUUGGCUGAUAU----UGACACCCUACAAACUGUGGGGU --....((-(...)))..((((..((((((((.....((....))....))))))))))))....((((..((((((......))))))..----))))(((((((.....))))))) ( -46.20) >DroEre_CAF1 12340 116 - 1 --ACUACAAUGGCCUGGCGGCGACGCUCUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCGGUUUGUCGUCGUCAGCAAUUUGGCUGAUAUAUAUGUACACCCUACAAACUGUGGGGU --..(((((((((..(((.(((..((((((((.....((....))....))))))))))).))).))))).((((((......)))))).....)))).(((((((.....))))))) ( -46.30) >DroYak_CAF1 12829 114 - 1 CAACUACAAGGGCCUGGCGGCGACGCUCUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCAUUUUGUCGUCGUCAGCAAUUUGGCUGAUAU----UGACACCCUACAAACUGUGGGGU ......((((...(((((((((((((((((((.....((....))....))))))))........)))))))))))...))))........----....(((((((.....))))))) ( -45.40) >consensus __ACUACAAGGGCCUGGCGGCGACGCUCUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAAUUUGGCUGAUAU____UGACACCCUACAAACUGUGGGGU ......((((...(((((((((((((((((((.....((....))....))))))))........)))))))))))...))))................(((((((.....))))))) (-44.08 = -44.68 + 0.60)


| Location | 9,430,441 – 9,430,549 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.64 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -23.17 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9430441 108 + 22224390 AUUGCUGACGACGACAAAAGGCGGCUCUCUCAUAAGCGGAAGCAAAAAGAGAGAGCGUCGCCGCCAGGCCCUUGUAGU------------UCCGCGUUUAUGGCACGAUAAGAACGAGAU ..((((((((..(((..((((..((((((((....((....)).....))))))))...(((....)))))))...))------------)...))))...))))............... ( -34.70) >DroSec_CAF1 12203 107 + 1 AUUGCUGACGACGACAAAAGGCGGCUCUCUCAUAAGCGGAAGCAAAAAGAGAGAGCGUCGCCGCCAGGCCC-UGUAGU------------UCCGCGUUUAUGGCACGAUAAGAAUGAGAU ..((((((((..(((....((((.(((((((....((....)).....)))))))))))(((....)))..-....))------------)...))))...))))............... ( -34.30) >DroSim_CAF1 12918 107 + 1 AUUGCUGACGACGACAAAAGGCGGCUCUCUCAUAAGCGGAAGCAAAAAGAGAGAGCGUCGCCGCCAGGCCC-UGUAGU------------UCUGCGUUUAUGGCACGAUAAGAAUGAGAU ..((((((((..(((....((((.(((((((....((....)).....)))))))))))(((....)))..-....))------------)...))))...))))............... ( -34.30) >DroEre_CAF1 12378 108 + 1 AUUGCUGACGACGACAAACCGCGGCUCUCUCAUAAGCGGAAGCAAAAAGAGAGAGCGUCGCCGCCAGGCCAUUGUAGU------------UCCGCGUUUAUGGCGUGAUAAGAACGAGAU ..(((.((((.((......))).((((((((....((....)).....)))))))))))(((....)))....)))((------------(((((((.....)))))....))))..... ( -35.10) >DroYak_CAF1 12863 120 + 1 AUUGCUGACGACGACAAAAUGCGGCUCUCUCAUAAGCGGAAGCAAAAAGAGAGAGCGUCGCCGCCAGGCCCUUGUAGUUGUAGUUGUUCUUCCGCGUGUAUGGCAUGAUAAGAACGAGAU ...((((.((((.((((......((((((((....((....)).....))))))))...(((....)))..)))).))))))))(((((((...((((.....))))..))))))).... ( -42.00) >DroAna_CAF1 9620 86 + 1 -AUGCUGACGAAAGCAAAUAUGAACUCUCU--UGAGCUC----------GGAGAGC---CCUUUUGGGGCC-UGUCGU------------UCA-----UGUAGGACAGUAAUGAUGAUAU -.(((((.(....)...(((((((((((((--.......----------)))))((---((.....)))).-....))------------)))-----)))....))))).......... ( -25.10) >consensus AUUGCUGACGACGACAAAAGGCGGCUCUCUCAUAAGCGGAAGCAAAAAGAGAGAGCGUCGCCGCCAGGCCC_UGUAGU____________UCCGCGUUUAUGGCACGAUAAGAACGAGAU ..((((...((((..........((((((((....((....)).....))))))))...(((....))).........................))))...))))............... (-23.17 = -23.88 + 0.71)



| Location | 9,430,441 – 9,430,549 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.64 |
| Mean single sequence MFE | -34.94 |
| Consensus MFE | -25.13 |
| Energy contribution | -26.28 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9430441 108 - 22224390 AUCUCGUUCUUAUCGUGCCAUAAACGCGGA------------ACUACAAGGGCCUGGCGGCGACGCUCUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAAU .....(((((...(((.......))).)))------------)).......((.((((((((((((((((((.....((....))....))))))))........))))))))))))... ( -38.60) >DroSec_CAF1 12203 107 - 1 AUCUCAUUCUUAUCGUGCCAUAAACGCGGA------------ACUACA-GGGCCUGGCGGCGACGCUCUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAAU ............(((((.......))))).------------......-..((.((((((((((((((((((.....((....))....))))))))........))))))))))))... ( -36.40) >DroSim_CAF1 12918 107 - 1 AUCUCAUUCUUAUCGUGCCAUAAACGCAGA------------ACUACA-GGGCCUGGCGGCGACGCUCUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAAU ......((((...(((.......))).)))------------).....-..((.((((((((((((((((((.....((....))....))))))))........))))))))))))... ( -35.20) >DroEre_CAF1 12378 108 - 1 AUCUCGUUCUUAUCACGCCAUAAACGCGGA------------ACUACAAUGGCCUGGCGGCGACGCUCUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCGGUUUGUCGUCGUCAGCAAU ................(((((.........------------......)))))(((((((((((((((((((.....((....))....))))))))........))))))))))).... ( -36.76) >DroYak_CAF1 12863 120 - 1 AUCUCGUUCUUAUCAUGCCAUACACGCGGAAGAACAACUACAACUACAAGGGCCUGGCGGCGACGCUCUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCAUUUUGUCGUCGUCAGCAAU .....((((((....(((.......))).))))))................((.((((((((((((((((((.....((....))....))))))))........))))))))))))... ( -40.60) >DroAna_CAF1 9620 86 - 1 AUAUCAUCAUUACUGUCCUACA-----UGA------------ACGACA-GGCCCCAAAAGG---GCUCUCC----------GAGCUCA--AGAGAGUUCAUAUUUGCUUUCGUCAGCAU- ............(((((.....-----...------------..))))-)((((.....))---)).....----------((((((.--...)))))).....((((......)))).- ( -22.10) >consensus AUCUCAUUCUUAUCGUGCCAUAAACGCGGA____________ACUACA_GGGCCUGGCGGCGACGCUCUCUCUUUUUGCUUCCGCUUAUGAGAGAGCCGCCUUUUGUCGUCGUCAGCAAU .....................................................(((((((((((((((((((.....((....))....))))))))........))))))))))).... (-25.13 = -26.28 + 1.15)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:24 2006