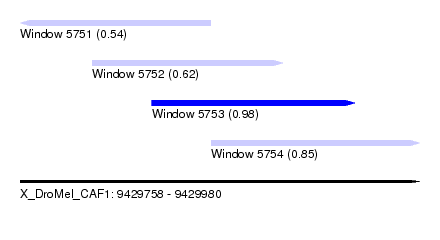
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,429,758 – 9,429,980 |
| Length | 222 |
| Max. P | 0.980605 |

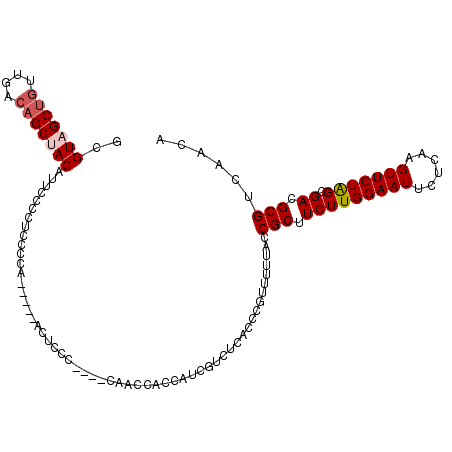
| Location | 9,429,758 – 9,429,864 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.41 |
| Mean single sequence MFE | -20.66 |
| Consensus MFE | -17.58 |
| Energy contribution | -18.14 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9429758 106 - 22224390 GCGUAGCUGUUGACAGCUACCUUCCCCUCCCCA-----ACUCCC----CUACCAUCAUCGUCUCACCCGUUUUUACCGCUUCUUGGAAUUCUCAAGUUCUAGCGACGCGUCAACA (((((((((....))))))).............-----......----...........(((......((....)).(((.(((((.....)))))....))))))))....... ( -21.50) >DroSec_CAF1 11514 106 - 1 ACGUAGCUAUUGACCGCUACAUUCCCCUCCCCA-----ACUCCG----CAACCACCAUCGUCUCACCCGUUUUUACCGCUUCUUGGAAUUCUCAAGUUCUAGCGACGCGUCAACA ..(((((........))))).............-----....((----(...................((....))((((.(((((.....)))))....))))..)))...... ( -17.60) >DroSim_CAF1 12227 106 - 1 ACGUAGCUGUUGACAGCUACAUUCCCCUCCCCA-----ACUCCG----CAACCACCUUCGUCUCACCCGUUUUUACCGCUUCUUGGAAUUCUCAAGUUCUGGCGACGCGUCAACA ..(((((((....))))))).............-----....((----(...................((....))((((.(((((.....)))))....))))..)))...... ( -22.30) >DroEre_CAF1 11649 115 - 1 GUGUAGCUGUUGGCAGCCACAUUUCCCUCUCCACCCCAACCCCCUCCACCACCAUCAUCGCCUCACCCGUUUUUACCGCUUCUUGGAAUUCUCAAGUUCUAGCGACGCGUCAACA ((((.((((....)))).))))....................................(((.((....((....)).(((.(((((.....)))))....))))).)))...... ( -18.50) >DroYak_CAF1 12145 106 - 1 GCGUAGCUGUUGGCAGCCACAUACCCCUCGCCA-----ACGCCC----CCACCACAUUCGCCUUACCCGUUUUUACCGCUUCUUGGAAUUCUCAAGUUCUGGCGACGCGUCAACA ((((.((.((((((((..........)).))))-----))))..----............................((((.(((((.....)))))....))))))))....... ( -23.40) >consensus GCGUAGCUGUUGACAGCUACAUUCCCCUCCCCA_____ACUCCC____CAACCACCAUCGUCUCACCCGUUUUUACCGCUUCUUGGAAUUCUCAAGUUCUAGCGACGCGUCAACA ..(((((((....)))))))........................................................(((.(((((((((......))))))).)).)))...... (-17.58 = -18.14 + 0.56)


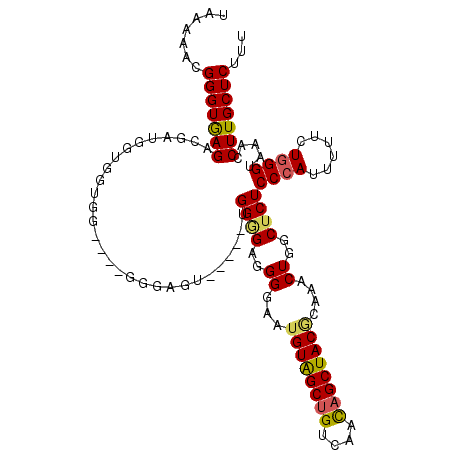
| Location | 9,429,798 – 9,429,904 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 86.13 |
| Mean single sequence MFE | -36.54 |
| Consensus MFE | -24.70 |
| Energy contribution | -24.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9429798 106 + 22224390 UAAAAACGGGUGAGACGAUGAUGGUAG----GGGAGU-----UGGGGAGGGGAAGGUAGCUGUCAACAGCUACGCAAACUUGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUU .......(((..(...........(((----..((((-----..(((((((.((((((((((....))))))).....))).)))))))))))..)))((....)))..)))... ( -36.90) >DroSec_CAF1 11554 106 + 1 UAAAAACGGGUGAGACGAUGGUGGUUG----CGGAGU-----UGGGGAGGGGAAUGUAGCGGUCAAUAGCUACGUAAACUAGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUU ...........(((.(((.(((...(.----(((((.-----..(((((((..(((((((........))))))).......)))))))....))))).)...)))))))))... ( -33.60) >DroSim_CAF1 12267 106 + 1 UAAAAACGGGUGAGACGAAGGUGGUUG----CGGAGU-----UGGGGAGGGGAAUGUAGCUGUCAACAGCUACGUAAACUGGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUU ...........(((.(.(((((...(.----(((((.-----..(((((((..(((((((((....))))))))).......)))))))....))))).)...)))))))))... ( -37.80) >DroEre_CAF1 11689 115 + 1 UAAAAACGGGUGAGGCGAUGAUGGUGGUGGAGGGGGUUGGGGUGGAGAGGGAAAUGUGGCUGCCAACAGCUACACAAACUGGCUCUCCCAAUUUUCUAGGUAAACCUUGCUCUUU .......(((..(((......(((...(((.((((((..(..((..........((((((((....))))))))))..)..))))))))).....)))......)))..)))... ( -39.40) >DroYak_CAF1 12185 106 + 1 UAAAAACGGGUAAGGCGAAUGUGGUGG----GGGCGU-----UGGCGAGGGGUAUGUGGCUGCCAACAGCUACGCAAACUUACUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUU .......((((((((.......(((((----(.((((-----.(((....((((......))))....)))))))...))))))..((((......))))....))))))))... ( -35.00) >consensus UAAAAACGGGUGAGACGAUGGUGGUGG____GGGAGU_____UGGGGAGGGGAAUGUAGCUGUCAACAGCUACGCAAACUGGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUU .......(((((((.............................((((..((...((((((((....))))))))....))..))))((((......)))).....)))))))... (-24.70 = -24.70 + -0.00)


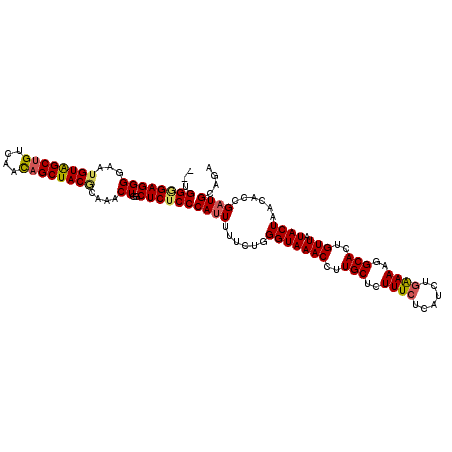
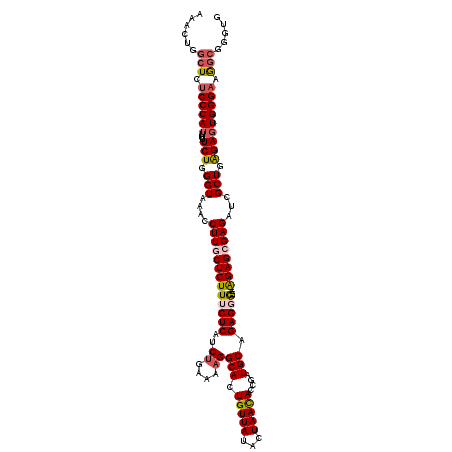
| Location | 9,429,831 – 9,429,944 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -28.46 |
| Energy contribution | -28.58 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9429831 113 + 22224390 --UGGGGAGGGGAAGGUAGCUGUCAACAGCUACGCAAACUUGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAUCUAAAACGGCAUUGUUAUACUAACACCGAUGCCGA --((((((((((((((((((((....)))))))(((....)))...((((......)))).....))).)))))))))).......((((((((............)))))))). ( -41.20) >DroSec_CAF1 11587 113 + 1 --UGGGGAGGGGAAUGUAGCGGUCAAUAGCUACGUAAACUAGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAUCUGAAAAGGCACUGUUAUACUAACACCGAUGCAGA --..(((((((..(((((((........))))))).......))))))).....((((.((......(((..((((......))))..))).(((((...)))))...)).)))) ( -31.60) >DroSim_CAF1 12300 113 + 1 --UGGGGAGGGGAAUGUAGCUGUCAACAGCUACGUAAACUGGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAUCUGAAAAGGCACUGUUAUACUAACACCGAUGCAGA --((((((((((.(((((((((....))))))))).....((....((((......))))....))...))))))))))((((....((...(((((...)))))))....)))) ( -37.10) >DroEre_CAF1 11729 115 + 1 GGUGGAGAGGGAAAUGUGGCUGCCAACAGCUACACAAACUGGCUCUCCCAAUUUUCUAGGUAAACCUUGCUCUUUCUCAUCUGGAAAGGCACUGUUAUACUAAUACCGAUGCAGA (((((((((((...((((((((....))))))))....))..))))))..........(((((((..(((.((((((.....)))))))))..))).))))...)))........ ( -34.30) >DroYak_CAF1 12218 113 + 1 --UGGCGAGGGGUAUGUGGCUGCCAACAGCUACGCAAACUUACUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAACUGGAAAGGCACUGUUAUACUAAUACCGAUGCAGA --.(((((((..((.(((((((....))))))).............((((......))))))..)))))))((((((.....))))))(((.((.(((....))).)).)))... ( -33.20) >consensus __UGGGGAGGGGAAUGUAGCUGUCAACAGCUACGCAAACUGGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAUCUGAAAAGGCACUGUUAUACUAACACCGAUGCAGA ...((((((((...((((((((....))))))))....))..))))))((((......(((((((..(((..((((......))))..)))..))).))))......)))).... (-28.46 = -28.58 + 0.12)


| Location | 9,429,864 – 9,429,980 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.23 |
| Mean single sequence MFE | -43.58 |
| Consensus MFE | -29.08 |
| Energy contribution | -30.16 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9429864 116 + 22224390 AAACUUGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAUCUAAAACGGCAUUGUUAUACUAACACCGAUGCCGAGGG----AGAGCGAGAUCGCUGAGAGUGGGAAAGCGGGUG ..(((((((.((((((...(((.(((....((((((((..(((.........(((((((............))))))))))..----))))))))...))).))))))))).))))))). ( -49.80) >DroSec_CAF1 11620 116 + 1 AAACUAGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAUCUGAAAAGGCACUGUUAUACUAACACCGAUGCAGAGGA----AGAGCGAGAUCGCUGAGAGUGGGAUGGCGGUUG .((((.(((.((((((...(((.(((....(((((((((((((..((....))(((.(((((...)))))....))).)))))----))))))))...))).))))))))).))))))). ( -45.40) >DroSim_CAF1 12333 116 + 1 AAACUGGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAUCUGAAAAGGCACUGUUAUACUAACACCGAUGCAGAGGA----AGAGCGAGAUCGCUGAGAGUGGGAAGGCGGUUG .(((((.((.((((((...(((.(((....(((((((((((((..((....))(((.(((((...)))))....))).)))))----))))))))...))).))))))))))).))))). ( -46.70) >DroEre_CAF1 11764 120 + 1 AAACUGGCUCUCCCAAUUUUCUAGGUAAACCUUGCUCUUUCUCAUCUGGAAAGGCACUGUUAUACUAAUACCGAUGCAGAGUGAGCGGGAGAGAGAUCGCUGAGAGUGGGAAGGCGGGUG ..(((.((..(((((.(((((.(((....))).((((((((((.(((....)))..(((((.((((.............)))))))))))))))))..)).))))))))))..)).))). ( -39.62) >DroYak_CAF1 12251 120 + 1 AAACUUACUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAACUGGAAAGGCACUGUUAUACUAAUACCGAUGCAGAGUGCGAGAGAGAGAGAUCGCUGGGAGUGGGUAGACGGCUG ..((((((((.((((......))))....((...(((((((((..((....))(((((.((((..........))).).)))))..)))))))))......))))))))))......... ( -36.40) >consensus AAACUGGCUCUCCCAUUUUUCUGGGUAAACCUUGCUCUUUCUCAUCUGAAAAGGCACUGUUAUACUAACACCGAUGCAGAGGG____AGAGCGAGAUCGCUGAGAGUGGGAAGGCGGGUG ......(((.((((((...(((.(((....((((((((......((((....((...(((((...)))))))....)))).......))))))))...))).))))))))).)))..... (-29.08 = -30.16 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:18 2006