| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,082,940 – 1,083,060 |
| Length | 120 |
| Max. P | 0.804598 |

| Location | 1,082,940 – 1,083,060 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -48.48 |
| Consensus MFE | -30.94 |
| Energy contribution | -31.33 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
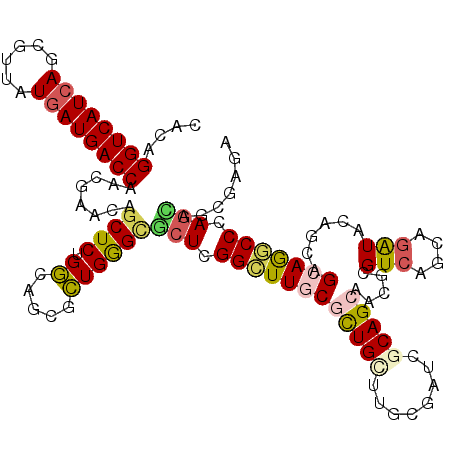
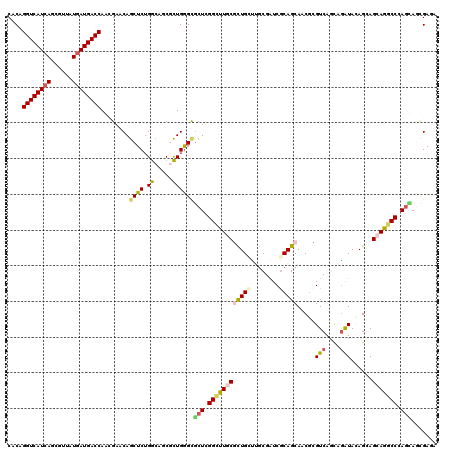
>X_DroMel_CAF1 1082940 120 + 22224390 CGCAGGUCAUCAGCGUAAUGAUGACCAAGGAGCAGCUCUGGCAGCGCUGGGCGCUCGGCUUCCUCUGGCGGCGAUCGCAGCAUCUGGUCAGCAGGUAGAGCAGCAGGCCCAGUAGGGAGC ....((((((((......))))))))...((((.((.(..((...))..))))))).(((((((((((.(.(....((.(((((((.....)))))...)).)).).))))).))))))) ( -50.30) >DroVir_CAF1 17030 120 + 1 CACAGGUCAUCAGCGUUAUGAUGACCAGCGAACAGCUCUGGCAGCGCUGGGUGCUCGGCUUGCGCUGCUUGCGAUCGCAGCAGCGCGUCAACAGAUACAGGAGCAGGCCGAGCAGCGAGA ....((((((((......))))))))((((....((....))..)))).(.((((((((((((((((((.((....))))))))(..((....))..)....)))))))))))).).... ( -60.60) >DroGri_CAF1 14396 120 + 1 CACAGGUCAUCAACGUUAUAAUGACCAGUGAGCAGCUCUGGCAACGCUGGGCGCUCGGCUUGCGCUGCUUGCGAUCGCAGCAACGCGUCAACAAAUACAGUAGCAGGCCAAGCAGCGAGA (((.((((((..........)))))).))).((.((.(..((...))..)))(((.((((((((((((........))))).((..((........)).)).))))))).))).)).... ( -44.50) >DroWil_CAF1 13751 120 + 1 CACAGGUCAUCAGGGUUAUGAUGACCAAGGAGCAACUCUGACACCUUUGGGCACUUGGUUUGCGUUGUUUUCGAUCGCAGCAUCUUGUCAGUAGAUAUAGUAGCAGACCCAAUAAUGAUA ....((((((((......))))))))((((.(((....)).).)))).........((((((((((((........)))))((..((((....))))..)).)))))))........... ( -32.20) >DroMoj_CAF1 24125 120 + 1 CACAGGUCAUCAGCGUUAUGAUGACCAGCGAACAGCUCUGGCAGCGCUGAGCGCUCGGCUUGCGCUGCUUGCGAUCGCAGCAACGCGUCAGUAGAUACAGCAGCAAGCCGAGCAACGAGA ....((((((((......))))))))..((....((((.(((...)))))))(((((((((((((((.((((...(((......)))...))))...)))).)))))))))))..))... ( -57.20) >DroAna_CAF1 15328 120 + 1 CACAGGUCAUCAGGGUGAUGAUGACCAACGAACAGCUCUGGCACCUCUGGGCGCUGGGCUUCCUCUGGCGGCGGUCACAGCAGCGGGUCAGGAGGUAGAGGAGCAGUCCCAGGAGCGAAA ....((((((((......))))))))..((....(((((...((((((.(((((((((((.((......)).)))).)))).....))).))))))...)))))..((....)).))... ( -46.10) >consensus CACAGGUCAUCAGCGUUAUGAUGACCAACGAACAGCUCUGGCAGCGCUGGGCGCUCGGCUUGCGCUGCUUGCGAUCGCAGCAACGCGUCAGCAGAUACAGCAGCAGGCCCAGCAGCGAGA ....((((((((......))))))))........((((.((.....))))))(((.((((((((((((........))))).....(((....)))......))))))).)))....... (-30.94 = -31.33 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:23 2006