
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,416,485 – 9,416,579 |
| Length | 94 |
| Max. P | 0.993769 |

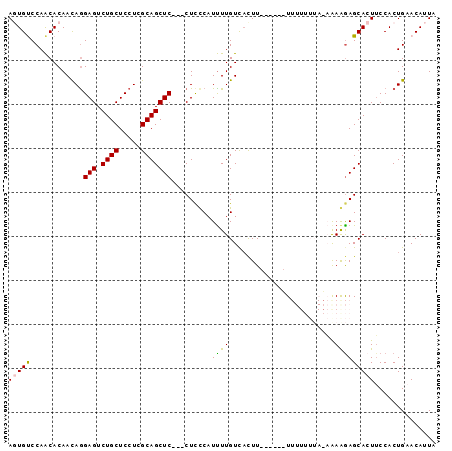
| Location | 9,416,485 – 9,416,579 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -19.12 |
| Consensus MFE | -8.52 |
| Energy contribution | -8.12 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
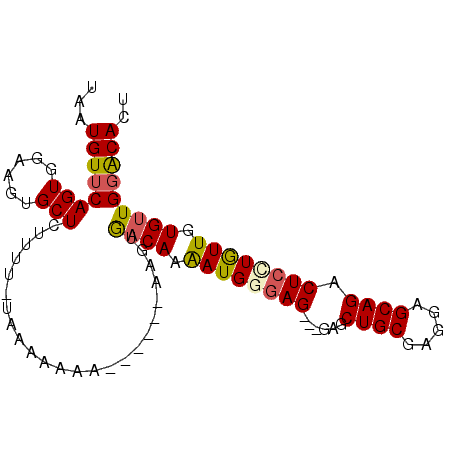
>X_DroMel_CAF1 9416485 94 + 22224390 AGUGUCCAACACAAUAGGAGUCUGCUCCUCGCAGCUC---CUUCCAUUUUGUCACUUUUUUUGUUUUUUUACAGAAGAGCACUUCCACUGAACAUUA (((((.(((......(((((.((((.....)))))))---))......))).....((((((((......))))))))))))).............. ( -22.10) >DroSec_CAF1 1815 92 + 1 AGUGUCCAACACAACAGGAGUCUGCUCCUCGCAGCUC---CUCCCAUUUUGUUUUUU--UUUUUUUUUUUAGAGAGGAGCACUUCCACUGAACAUUA (((((.((.......(((((.((((.....)))))))---)).......((((((((--(((........))))))))))).......)).))))). ( -22.90) >DroSim_CAF1 1527 88 + 1 AGUGUCCAACACAACAAGAGUCUGCUCCUCGCAGCUC---CUCCCAUUUUGUCACUU------UUUUUUUAGAAAGGAGCACUUCCACUGAACAUUA (((((.((.........(((.((((.....)))))))---.........(((..(((------(((....))))))..))).......)).))))). ( -17.90) >DroEre_CAF1 1746 80 + 1 ACUGCCCAACACAACAGGAGUCUGCUCCUCGCAGCUC---CUCUCAUUUUGUCACUU------U--------CAAAAAGCAAUUCCACUGCACAUUA ..(((..........(((((.((((.....)))))))---))....(((((......------.--------))))).)))................ ( -16.10) >DroYak_CAF1 1737 83 + 1 ACUGCCCAGCACAACAGGAGUCUGCUCCUCGCAGCUCCUCCUCUCAUCUUGUCACUU------U--------CAAAGAGCAAUGCCACUGAACAUUG ...((...((.....(((((.((((.....))))))))).......((((.......------.--------..))))))...))............ ( -16.60) >consensus AGUGUCCAACACAACAGGAGUCUGCUCCUCGCAGCUC___CUCCCAUUUUGUCACUU______UUUUUUUA_AAAAGAGCACUUCCACUGAACAUUA (((((............(((.((((.....))))))).........((((........................))))))))).............. ( -8.52 = -8.12 + -0.40)


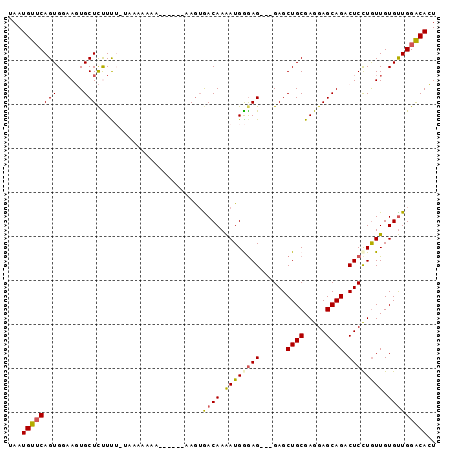
| Location | 9,416,485 – 9,416,579 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -26.44 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9416485 94 - 22224390 UAAUGUUCAGUGGAAGUGCUCUUCUGUAAAAAAACAAAAAAAGUGACAAAAUGGAAG---GAGCUGCGAGGAGCAGACUCCUAUUGUGUUGGACACU ...((((..(..((((....))))..).....)))).....((((.(((.(..(.((---(((((((.....)))).))))).)..).)))..)))) ( -26.10) >DroSec_CAF1 1815 92 - 1 UAAUGUUCAGUGGAAGUGCUCCUCUCUAAAAAAAAAAA--AAAAAACAAAAUGGGAG---GAGCUGCGAGGAGCAGACUCCUGUUGUGUUGGACACU ...((((((..(.(((((((((((((............--............)))))---)))).)).(((((....))))).)).)..)))))).. ( -28.76) >DroSim_CAF1 1527 88 - 1 UAAUGUUCAGUGGAAGUGCUCCUUUCUAAAAAAA------AAGUGACAAAAUGGGAG---GAGCUGCGAGGAGCAGACUCUUGUUGUGUUGGACACU ...(((((...(((((.....)))))........------....((((.((..((((---...((((.....)))).))))..)).))))))))).. ( -24.90) >DroEre_CAF1 1746 80 - 1 UAAUGUGCAGUGGAAUUGCUUUUUG--------A------AAGUGACAAAAUGAGAG---GAGCUGCGAGGAGCAGACUCCUGUUGUGUUGGGCAGU .....(((.......(..(((....--------.------)))..)(((.(..(.((---(((((((.....)))).))))).)..).))).))).. ( -24.70) >DroYak_CAF1 1737 83 - 1 CAAUGUUCAGUGGCAUUGCUCUUUG--------A------AAGUGACAAGAUGAGAGGAGGAGCUGCGAGGAGCAGACUCCUGUUGUGCUGGGCAGU ...(((((((((((....(((((..--------.------............))))).(((((((((.....)))).))))))))..)))))))).. ( -27.76) >consensus UAAUGUUCAGUGGAAGUGCUCUUUU_UAAAAAAA______AAGUGACAAAAUGGGAG___GAGCUGCGAGGAGCAGACUCCUGUUGUGUUGGACACU ...((((((((......)))........................((((.((((((((......((((.....)))).)))))))).))))))))).. (-16.30 = -16.50 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:05 2006