
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,394,635 – 9,394,770 |
| Length | 135 |
| Max. P | 0.993988 |

| Location | 9,394,635 – 9,394,755 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -28.31 |
| Energy contribution | -28.12 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9394635 120 + 22224390 ACAUGAUUUAUAAGUCGAAUUAGAAGGAGCCACACGACGGGCAUGGAACAAGAGUGAAUUAAUUACGAAUUCAAGCCCGAAGAGGUAAUUAAUUCACUUCAUUUUCUUCCCCUGCAAAGU ...(((((....))))).....(((((((((........))).........(((((((((((((((...(((......)))...)))))))))))))))....))))))........... ( -29.00) >DroSec_CAF1 8553 120 + 1 ACAUGAUUUAUAAGUCGAAUUAGAAGGAGCCACACGACGGGCAUGGAACAAGAGUGAAUUAAUUACGAAUUCAAGCCCGAAGAGGUAAUUAAUUCACUUCAUUUUCUUCCCCAGCAAAGU .............((((.................))))(((...((((...(((((((((((((((...(((......)))...)))))))))))))))....))))..)))........ ( -29.03) >DroSim_CAF1 12743 120 + 1 ACAUGAUUUAUAAGUCGAAUUAGAAGGAGCCACACGACGGGCAUGGAACAAGAGUGAAUUAAUUACGAAUUCAAGCCCGAAGAGGUAAUUAAUUCACUUCAUUUUCUUCCCCAGCAAAGU .............((((.................))))(((...((((...(((((((((((((((...(((......)))...)))))))))))))))....))))..)))........ ( -29.03) >DroYak_CAF1 12782 110 + 1 ACAUGAUUUAUAAGUCGAAUUAGAAGGAGCCACACGACGGGCAUGGAACAAGAGUGAAUUAAUUACGAAUUCAAGCCCGAAGAGGUAAUUAAUUCACUUCAUUUUCCUCC---------- ...(((((....)))))........((((((........)))..((((...(((((((((((((((...(((......)))...)))))))))))))))....)))))))---------- ( -29.40) >consensus ACAUGAUUUAUAAGUCGAAUUAGAAGGAGCCACACGACGGGCAUGGAACAAGAGUGAAUUAAUUACGAAUUCAAGCCCGAAGAGGUAAUUAAUUCACUUCAUUUUCUUCCCCAGCAAAGU ...(((((....)))))........((((((........)))..((((...(((((((((((((((...(((......)))...)))))))))))))))....))))))).......... (-28.31 = -28.12 + -0.19)



| Location | 9,394,635 – 9,394,755 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -27.08 |
| Energy contribution | -27.32 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9394635 120 - 22224390 ACUUUGCAGGGGAAGAAAAUGAAGUGAAUUAAUUACCUCUUCGGGCUUGAAUUCGUAAUUAAUUCACUCUUGUUCCAUGCCCGUCGUGUGGCUCCUUCUAAUUCGACUUAUAAAUCAUGU .....(((..((((.((...((.(((((((((((((...((((....))))...))))))))))))))))).)))).)))..((((..(((......)))...))))............. ( -29.20) >DroSec_CAF1 8553 120 - 1 ACUUUGCUGGGGAAGAAAAUGAAGUGAAUUAAUUACCUCUUCGGGCUUGAAUUCGUAAUUAAUUCACUCUUGUUCCAUGCCCGUCGUGUGGCUCCUUCUAAUUCGACUUAUAAAUCAUGU .........(((..(((...((.(((((((((((((...((((....))))...)))))))))))))))...)))....)))((((..(((......)))...))))............. ( -28.70) >DroSim_CAF1 12743 120 - 1 ACUUUGCUGGGGAAGAAAAUGAAGUGAAUUAAUUACCUCUUCGGGCUUGAAUUCGUAAUUAAUUCACUCUUGUUCCAUGCCCGUCGUGUGGCUCCUUCUAAUUCGACUUAUAAAUCAUGU .........(((..(((...((.(((((((((((((...((((....))))...)))))))))))))))...)))....)))((((..(((......)))...))))............. ( -28.70) >DroYak_CAF1 12782 110 - 1 ----------GGAGGAAAAUGAAGUGAAUUAAUUACCUCUUCGGGCUUGAAUUCGUAAUUAAUUCACUCUUGUUCCAUGCCCGUCGUGUGGCUCCUUCUAAUUCGACUUAUAAAUCAUGU ----------((((((.((..(((((((((((((((...((((....))))...)))))))))))))).)..))((((((.....)))))).))))))...................... ( -28.60) >consensus ACUUUGCUGGGGAAGAAAAUGAAGUGAAUUAAUUACCUCUUCGGGCUUGAAUUCGUAAUUAAUUCACUCUUGUUCCAUGCCCGUCGUGUGGCUCCUUCUAAUUCGACUUAUAAAUCAUGU .........(((..(((...((.(((((((((((((...((((....))))...)))))))))))))))...)))....)))((((..(((......)))...))))............. (-27.08 = -27.32 + 0.25)


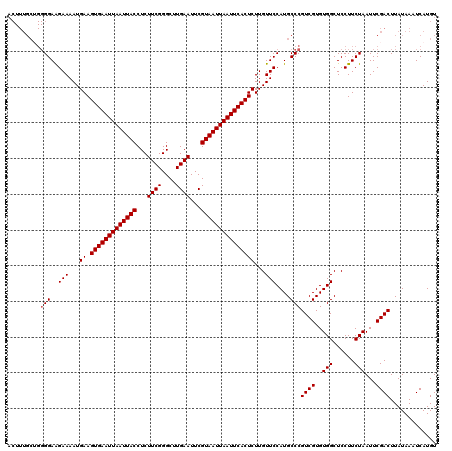
| Location | 9,394,675 – 9,394,770 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 83.16 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -20.69 |
| Energy contribution | -20.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.92 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9394675 95 + 22224390 GCAUGGAACAAGAGUGAAUUAAUUACGAAUUCAAGCCCGAAGAGGUAAUUAAUUCACUUCAUUUUCUUCCCCUGCAAAGUUGGCUGAAAACCAAA (((.((((.(((((((((((((((((...(((......)))...)))))))))))))))....)).))))..)))....((((.(....))))). ( -26.40) >DroSim_CAF1 12783 95 + 1 GCAUGGAACAAGAGUGAAUUAAUUACGAAUUCAAGCCCGAAGAGGUAAUUAAUUCACUUCAUUUUCUUCCCCAGCAAAGUUGACUGAAAACCAAA ...(((.....(((((((((((((((...(((......)))...)))))))))))))))..(((((.....((((...))))...)))))))).. ( -23.70) >DroYak_CAF1 12822 73 + 1 GCAUGGAACAAGAGUGAAUUAAUUACGAAUUCAAGCCCGAAGAGGUAAUUAAUUCACUUCAUUUUCCUCC----------------------AAA ....((((...(((((((((((((((...(((......)))...)))))))))))))))....))))...----------------------... ( -22.20) >consensus GCAUGGAACAAGAGUGAAUUAAUUACGAAUUCAAGCCCGAAGAGGUAAUUAAUUCACUUCAUUUUCUUCCCC_GCAAAGUUG_CUGAAAACCAAA ....((((...(((((((((((((((...(((......)))...)))))))))))))))....))))............................ (-20.69 = -20.47 + -0.22)


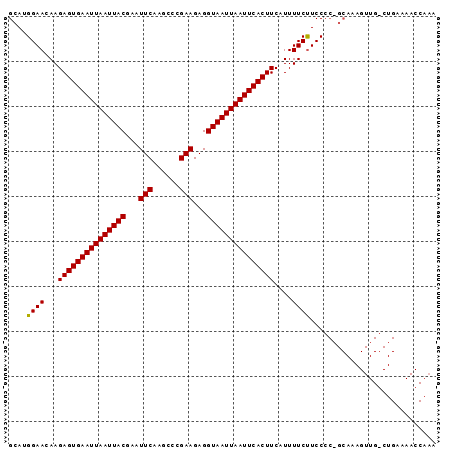
| Location | 9,394,675 – 9,394,770 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 83.16 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -20.42 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9394675 95 - 22224390 UUUGGUUUUCAGCCAACUUUGCAGGGGAAGAAAAUGAAGUGAAUUAAUUACCUCUUCGGGCUUGAAUUCGUAAUUAAUUCACUCUUGUUCCAUGC .(((((.....)))))....(((..((((.((...((.(((((((((((((...((((....))))...))))))))))))))))).)))).))) ( -26.90) >DroSim_CAF1 12783 95 - 1 UUUGGUUUUCAGUCAACUUUGCUGGGGAAGAAAAUGAAGUGAAUUAAUUACCUCUUCGGGCUUGAAUUCGUAAUUAAUUCACUCUUGUUCCAUGC ..(((((..((((.......))))..))....((..(((((((((((((((...((((....))))...)))))))))))))).)..)))))... ( -24.10) >DroYak_CAF1 12822 73 - 1 UUU----------------------GGAGGAAAAUGAAGUGAAUUAAUUACCUCUUCGGGCUUGAAUUCGUAAUUAAUUCACUCUUGUUCCAUGC ...----------------------...((((...((.(((((((((((((...((((....))))...)))))))))))))))...)))).... ( -22.00) >consensus UUUGGUUUUCAG_CAACUUUGC_GGGGAAGAAAAUGAAGUGAAUUAAUUACCUCUUCGGGCUUGAAUUCGUAAUUAAUUCACUCUUGUUCCAUGC .........................((((.((...((.(((((((((((((...((((....))))...))))))))))))))))).)))).... (-20.42 = -20.20 + -0.22)

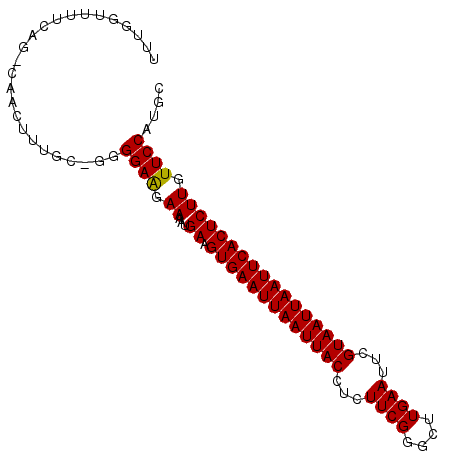
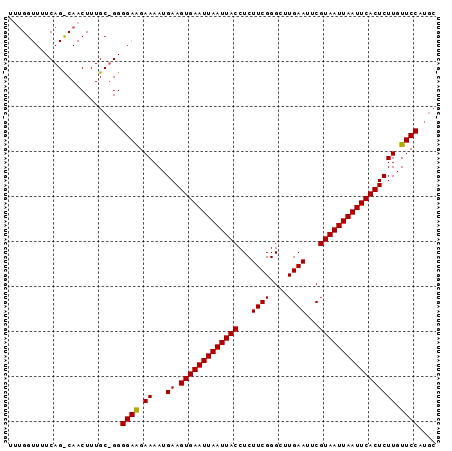
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:58 2006