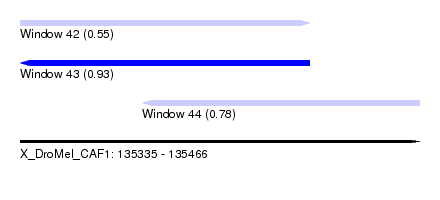
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 135,335 – 135,466 |
| Length | 131 |
| Max. P | 0.929147 |

| Location | 135,335 – 135,430 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 91.93 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -18.62 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 135335 95 + 22224390 UUGUAGGCUGGCCGUUGCCCUGUUCCAAUUGGAACUGUACUUUUAUAGCCCACCCCAAUACA----ACCUAUAACAGUGGCGUCCUUCACUAUUGGAAC ..((.(((((((....)))..(((((....)))))...........)))).)).(((((...----.........(((((......))))))))))... ( -20.80) >DroSec_CAF1 7500 95 + 1 UUGUAGGCUGGCCGUUGCCCUGUUCCAAUUGGAACUGUACUUUUAUAGCCCACCCCAAUACA----ACCUAUAACAGUGGCGUCUUUCACUAUUGGAAC ..((.(((((((....)))..(((((....)))))...........)))).)).(((((...----.........(((((......))))))))))... ( -20.80) >DroSim_CAF1 7279 95 + 1 UUGUAGGCUGGCCGUUGCCCUGUUCCAAUUGGAACUGUACUUUUAUAGCCCACCCCAAUAUA----ACCUAUAACAGUGGCGUCUUUCACUAUUGGAAC ..((.(((((((....)))..(((((....)))))...........)))).)).(((((...----.........(((((......))))))))))... ( -20.80) >DroEre_CAF1 6545 95 + 1 AUGUAGGGUGGCCCUUGCCCUGUUCCAAUUGGAACUGUACUUUUAUAGUCCACCCCAAUAAA----ACCUAUCACAGUGGCGUCCUUCACCAUUGGAAC .....((((((..........(((((....)))))...(((.....))))))))).......----.....((.((((((.(.....).)))))))).. ( -24.90) >DroYak_CAF1 6797 98 + 1 AUGUAGGGUGGCCCUUGUCCUGUUCCAAUUGGAACUGUACUUUCAUAG-CCACCCCAAUAGAACGAACCCAUCACAGUGGCGUCCUUCACUAUUGGAAC .....(((((((....((.(.(((((....))))).).)).......)-))))))((((((...(.((((((....)))).)).)....)))))).... ( -29.30) >consensus UUGUAGGCUGGCCGUUGCCCUGUUCCAAUUGGAACUGUACUUUUAUAGCCCACCCCAAUACA____ACCUAUAACAGUGGCGUCCUUCACUAUUGGAAC .((((((..(((....)))..(((((....)))))................................)))))).((((((.(.....).)))))).... (-18.62 = -18.90 + 0.28)

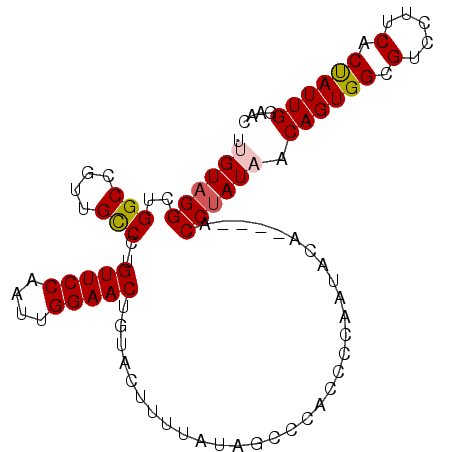
| Location | 135,335 – 135,430 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 91.93 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -23.56 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 135335 95 - 22224390 GUUCCAAUAGUGAAGGACGCCACUGUUAUAGGU----UGUAUUGGGGUGGGCUAUAAAAGUACAGUUCCAAUUGGAACAGGGCAACGGCCAGCCUACAA ..(((((((((.....))(((.........)))----..)))))))(((((((...........(((((....)))))..(((....)))))))))).. ( -29.40) >DroSec_CAF1 7500 95 - 1 GUUCCAAUAGUGAAAGACGCCACUGUUAUAGGU----UGUAUUGGGGUGGGCUAUAAAAGUACAGUUCCAAUUGGAACAGGGCAACGGCCAGCCUACAA ..(((((((((.....))(((.........)))----..)))))))(((((((...........(((((....)))))..(((....)))))))))).. ( -29.40) >DroSim_CAF1 7279 95 - 1 GUUCCAAUAGUGAAAGACGCCACUGUUAUAGGU----UAUAUUGGGGUGGGCUAUAAAAGUACAGUUCCAAUUGGAACAGGGCAACGGCCAGCCUACAA ..(((((((((.....))(((.........)))----..)))))))(((((((...........(((((....)))))..(((....)))))))))).. ( -29.50) >DroEre_CAF1 6545 95 - 1 GUUCCAAUGGUGAAGGACGCCACUGUGAUAGGU----UUUAUUGGGGUGGACUAUAAAAGUACAGUUCCAAUUGGAACAGGGCAAGGGCCACCCUACAU ((..((.(((((.....))))).))..))....----.....((((((((.((......((...(((((....)))))...))...))))))))))... ( -29.30) >DroYak_CAF1 6797 98 - 1 GUUCCAAUAGUGAAGGACGCCACUGUGAUGGGUUCGUUCUAUUGGGGUGG-CUAUGAAAGUACAGUUCCAAUUGGAACAGGACAAGGGCCACCCUACAU ....((((((....((((.(((......)))))))...))))))((((((-((......((...(((((....)))))...))...))))))))..... ( -35.80) >consensus GUUCCAAUAGUGAAGGACGCCACUGUUAUAGGU____UGUAUUGGGGUGGGCUAUAAAAGUACAGUUCCAAUUGGAACAGGGCAACGGCCAGCCUACAA ..(((((((.........(((.........)))......)))))))(((((((...........(((((....)))))..(((....)))))))))).. (-23.56 = -24.40 + 0.84)

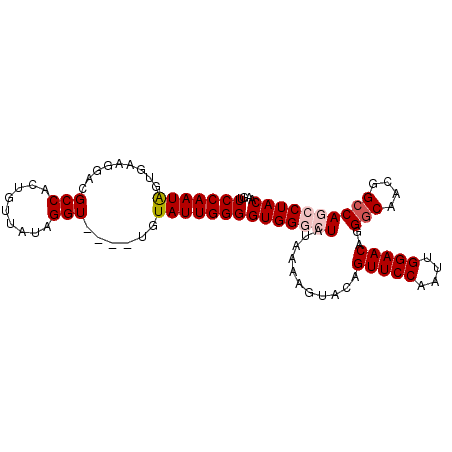
| Location | 135,375 – 135,466 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 91.43 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.22 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 135375 91 - 22224390 UGGCUAGCUCACAGCAACCUUCAUUGGGUUU--UUCCAGUUCCAAUAGUGAAGGACGCCACUGUUAUAGGU----UGUAUUGGGGUGGGCUAUAAAA (((((.((((.(((((((((..((((((...--.))))))...(((((((........)))))))..))))----)))..)))))).)))))..... ( -29.60) >DroSec_CAF1 7540 91 - 1 UGGCUAGCUCACAGCAACCUUCAUUGGGUUU--UUCCAGUUCCAAUAGUGAAAGACGCCACUGUUAUAGGU----UGUAUUGGGGUGGGCUAUAAAA (((((.((((.(((((((((..((((((...--.))))))...(((((((........)))))))..))))----)))..)))))).)))))..... ( -29.60) >DroSim_CAF1 7319 91 - 1 UGGCUAGCUCACAGCAACCUUCAUUGGGUUU--UUCCAGUUCCAAUAGUGAAAGACGCCACUGUUAUAGGU----UAUAUUGGGGUGGGCUAUAAAA ....((((((((...(((((.....))))).--.((((((.(((((((((........)))))))...)).----...))))))))))))))..... ( -27.60) >DroEre_CAF1 6585 93 - 1 UGGCUAGCUCGCAGCAACCUUCAUUGGGUUUUUUUUCAGUUCCAAUGGUGAAGGACGCCACUGUGAUAGGU----UUUAUUGGGGUGGACUAUAAAA ((((((.((((....(((((((((((((((((((.(((.......))).))))))).)))..)))).))))----)....)))).))).)))..... ( -24.70) >DroYak_CAF1 6837 94 - 1 UGGCUAGCUCGCAGCAACCUUCAAUGGGUUU--UUUCAGUUCCAAUAGUGAAGGACGCCACUGUGAUGGGUUCGUUCUAUUGGGGUGG-CUAUGAAA ..(((.(....))))..((((((.((((...--.......))))....))))))..((((((.(((((((.....))))))).)))))-)....... ( -29.80) >consensus UGGCUAGCUCACAGCAACCUUCAUUGGGUUU__UUCCAGUUCCAAUAGUGAAGGACGCCACUGUUAUAGGU____UGUAUUGGGGUGGGCUAUAAAA ....((((((((...(((((.....)))))....((((((.((.((((((........))))))....))........))))))))))))))..... (-21.46 = -21.22 + -0.24)

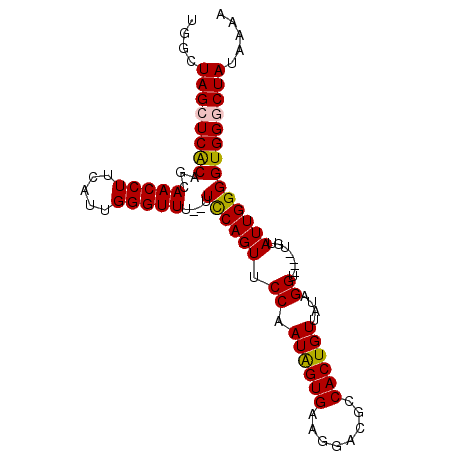
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:57 2006