
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,393,764 – 9,394,037 |
| Length | 273 |
| Max. P | 0.968223 |

| Location | 9,393,764 – 9,393,884 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.01 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -20.39 |
| Energy contribution | -20.07 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
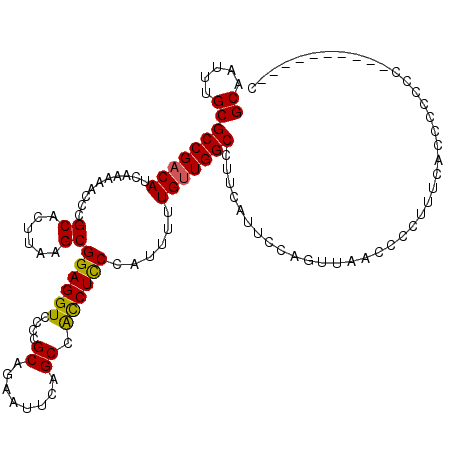
>X_DroMel_CAF1 9393764 120 - 22224390 GCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAGGUCCCCGCAGAAUUCAGCCACCUCCCAUUUUUGUUGGCCUUCAUUCCAGUUAACCCCUUUCACCCCUCCCCCCCUCCCCU ((.....))(((((((..........((......))((((((....((........)).))))))......))))))).......................................... ( -23.00) >DroSec_CAF1 7692 110 - 1 GCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAGGUCCCCGCAGAAUUCCGCCGUCUUCCAUUUUUGUUGGCCUUCAUUCCAGUUAACCCCUUUCACCCCCCG----------U ((.....))(((((((..........((......))(((((.(...((.(....).)).).))))).....)))))))...............................----------. ( -21.20) >DroSim_CAF1 11882 110 - 1 GCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAGGUCCCCGCAGAAUUCAGCCACCUCCCAUUUUUGCUGGCCUUCAUUCCAGUUAACCCCUUUCACCCCCCG----------C ((..((((((..(((.........((((......))))..)))..)))))).....................(((((........)))))..................)----------) ( -19.90) >DroYak_CAF1 12119 110 - 1 GCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAGGUCCCCGCAGAAUUCAGCCACCUCCCAUUUUUGUUGGCCUUCAUUCCAGUUAACCCCUUUCACCCACUC----------C ((.....))(((((((..........((......))((((((....((........)).))))))......)))))))...............................----------. ( -23.00) >consensus GCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAGGUCCCCGCAGAAUUCAGCCACCUCCCAUUUUUGUUGGCCUUCAUUCCAGUUAACCCCUUUCACCCCCCC__________C ((.....))(((((((..........((......))((((((....((........)).))))))......))))))).......................................... (-20.39 = -20.07 + -0.31)


| Location | 9,393,804 – 9,393,924 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -26.49 |
| Energy contribution | -26.17 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9393804 120 - 22224390 UUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUUGCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAGGUCCCCGCAGAAUUCAGCCACCUCCCAUUUUUGUUGGCCU ..........((...(((((((.((.........)).)))))))...))(((((((..........((......))((((((....((........)).))))))......))))))).. ( -29.10) >DroSec_CAF1 7722 120 - 1 UUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUUGCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAGGUCCCCGCAGAAUUCCGCCGUCUUCCAUUUUUGUUGGCCU ..........((...(((((((.((.........)).)))))))...))(((((((..........((......))(((((.(...((.(....).)).).))))).....))))))).. ( -27.30) >DroSim_CAF1 11912 120 - 1 UUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUUGCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAGGUCCCCGCAGAAUUCAGCCACCUCCCAUUUUUGCUGGCCU ..........((...(((((((.((.........)).)))))))...))((((....((((((...((......))((((((....((........)).))))))..)))))).)))).. ( -25.20) >DroYak_CAF1 12149 120 - 1 UUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUUGCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAGGUCCCCGCAGAAUUCAGCCACCUCCCAUUUUUGUUGGCCU ..........((...(((((((.((.........)).)))))))...))(((((((..........((......))((((((....((........)).))))))......))))))).. ( -29.10) >consensus UUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUUGCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAGGUCCCCGCAGAAUUCAGCCACCUCCCAUUUUUGUUGGCCU ..........((...(((((((.((.........)).)))))))...))(((((((..........((......))((((((....((........)).))))))......))))))).. (-26.49 = -26.17 + -0.31)


| Location | 9,393,844 – 9,393,964 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -31.94 |
| Energy contribution | -32.25 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9393844 120 - 22224390 CCCUAACCGGUUAGGGGAUGCAAAUAAUUUUCAAGCAAUUUUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUUGCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAG .(((((....)))))((.(((((((.........((((((......))))))....((((((.((.........)).)))))))))))))))............((((......)))).. ( -29.80) >DroSec_CAF1 7762 120 - 1 CCCUAUCCGGUUAGGGGGUGCAAAUAAUUUUCAAGCAAUUUUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUUGCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAG (((((......)))))(((((((((.........((((((......))))))....((((((.((.........)).)))))))))))))))............((((......)))).. ( -35.50) >DroSim_CAF1 11952 120 - 1 CCCUAUCCGGUUAGGGGGUGCAAAUAAUUUUCAAGCAAUUUUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUUGCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAG (((((......)))))(((((((((.........((((((......))))))....((((((.((.........)).)))))))))))))))............((((......)))).. ( -35.50) >DroYak_CAF1 12189 119 - 1 CUCUAUCGGGUU-GGGGGUGCAAAUAAUUUUCAAGCAAUUUUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUUGCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAG ((((...(((((-(..(((((((((.........((((((......))))))....((((((.((.........)).)))))))))))))))..))......))))((......)))))) ( -32.60) >consensus CCCUAUCCGGUUAGGGGGUGCAAAUAAUUUUCAAGCAAUUUUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUUGCAAUUUGCGCCGACAUCAAAAACCCGCACUUAAGCGGAG (((((......)))))(((((((((.........((((((......))))))....((((((.((.........)).)))))))))))))))............((((......)))).. (-31.94 = -32.25 + 0.31)


| Location | 9,393,884 – 9,394,004 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.98 |
| Mean single sequence MFE | -18.52 |
| Consensus MFE | -15.75 |
| Energy contribution | -16.25 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9393884 120 + 22224390 AAAGAAAAUGAUUUAUUAUCUGCAAAAGGCAAUUAAGUAAAAUUGCUUGAAAAUUAUUUGCAUCCCCUAACCGGUUAGGGAAAAAUUUCAAAUAGAAGCAAAACCAAAAUACACGCAAAU ....................(((...((((((((......))))))))........(((((...((((((....))))))......(((.....))))))))............)))... ( -23.10) >DroSec_CAF1 7802 119 + 1 AAAGAAAAUGAUUUAUUAUCUGCAAAAGGCAAUUAAGUAAAAUUGCUUGAAAAUUAUUUGCACCCCCUAACCGGAUAGGGAAAA-AGCCAAAUAGAAGCAAAACCAAAAUAUACGCAAAU .....(((((((((......(((.....))).(((((((....))))))))))))))))((...(((((......)))))....-.((.........))...............)).... ( -19.00) >DroSim_CAF1 11992 119 + 1 AAAGAAAAUGAUUUAUUAUCUGCAAAAGGCAAUUAAGUAAAAUUGCUUGAAAAUUAUUUGCACCCCCUAACCGGAUAGGGAAAA-UGCCAAAUAGAAGCAAAACCAAAAUAUACGCAAAU ....................(((...((((((((......)))))))).....(((((((((..(((((......)))))....-)).)))))))...................)))... ( -20.40) >DroYak_CAF1 12229 118 + 1 AAAGAAAAUGAUUUAUUAUCUGCAAAAGGCAAUUAAGUAAAAUUGCUUGAAAAUUAUUUGCACCCCC-AACCCGAUAGAGAAAA-AGCCAAACAGAAGCAAAACCAAAAUAAACGCAAAU ....................(((...((((((((......))))))))........(((((......-................-............)))))............)))... ( -11.58) >consensus AAAGAAAAUGAUUUAUUAUCUGCAAAAGGCAAUUAAGUAAAAUUGCUUGAAAAUUAUUUGCACCCCCUAACCGGAUAGGGAAAA_AGCCAAAUAGAAGCAAAACCAAAAUAUACGCAAAU ....................(((...((((((((......))))))))........(((((...(((((......)))))........(.....)..)))))............)))... (-15.75 = -16.25 + 0.50)


| Location | 9,393,884 – 9,394,004 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.98 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -19.91 |
| Energy contribution | -19.60 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9393884 120 - 22224390 AUUUGCGUGUAUUUUGGUUUUGCUUCUAUUUGAAAUUUUUCCCUAACCGGUUAGGGGAUGCAAAUAAUUUUCAAGCAAUUUUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUU (((((((.(((.((.(((.((((((.((((((......((((((((....))))))))..))))))......))))))....))).)).)))....)))))))................. ( -25.30) >DroSec_CAF1 7802 119 - 1 AUUUGCGUAUAUUUUGGUUUUGCUUCUAUUUGGCU-UUUUCCCUAUCCGGUUAGGGGGUGCAAAUAAUUUUCAAGCAAUUUUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUU ((((((((((...........(((.......))).-...((((((......)))))))))).............((((((......)))))).....))))))................. ( -22.80) >DroSim_CAF1 11992 119 - 1 AUUUGCGUAUAUUUUGGUUUUGCUUCUAUUUGGCA-UUUUCCCUAUCCGGUUAGGGGGUGCAAAUAAUUUUCAAGCAAUUUUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUU ((((((((((..........((((.......))))-...((((((......)))))))))).............((((((......)))))).....))))))................. ( -24.00) >DroYak_CAF1 12229 118 - 1 AUUUGCGUUUAUUUUGGUUUUGCUUCUGUUUGGCU-UUUUCUCUAUCGGGUU-GGGGGUGCAAAUAAUUUUCAAGCAAUUUUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUU ..............((((((....(((((..(((.-.((.........(.((-(..(((.......)))..))).)..........))..)))....)))))....))))))........ ( -21.41) >consensus AUUUGCGUAUAUUUUGGUUUUGCUUCUAUUUGGCA_UUUUCCCUAUCCGGUUAGGGGGUGCAAAUAAUUUUCAAGCAAUUUUACUUAAUUGCCUUUUGCAGAUAAUAAAUCAUUUUCUUU (((((((...........(((((.((.....)).....(((((((......))))))).)))))..........((((((......))))))....)))))))................. (-19.91 = -19.60 + -0.31)

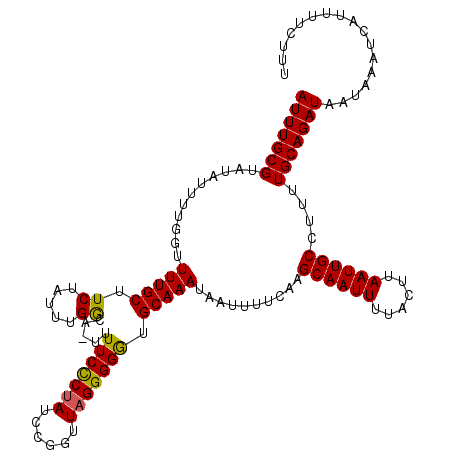
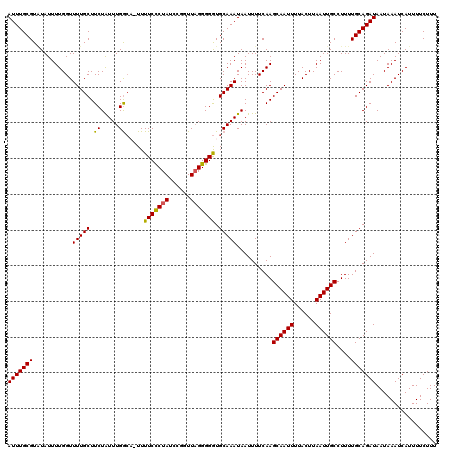
| Location | 9,393,924 – 9,394,037 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -17.60 |
| Consensus MFE | -12.84 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9393924 113 + 22224390 AAUUGCUUGAAAAUUAUUUGCAUCCCCUAACCGGUUAGGGAAAAAUUUCAAAUAGAAGCAAAACCAAAAUACACGCAAAUGACAGCAAAUCUUUUAGACAA-------UUCCAAAAAAUG ..(((((.(....((((((((...((((((....)))))).....((((.....))))................)))))))))))))).............-------............ ( -19.70) >DroSec_CAF1 7842 112 + 1 AAUUGCUUGAAAAUUAUUUGCACCCCCUAACCGGAUAGGGAAAA-AGCCAAAUAGAAGCAAAACCAAAAUAUACGCAAAUGACAGCAAAUCUCUCAGACAA-------UUUCAAAAAAUU ..(((((.(....((((((((...(((((......)))))....-.((.........))...............)))))))))))))).............-------............ ( -17.60) >DroSim_CAF1 12032 112 + 1 AAUUGCUUGAAAAUUAUUUGCACCCCCUAACCGGAUAGGGAAAA-UGCCAAAUAGAAGCAAAACCAAAAUAUACGCAAAUGACAGCAAAUCUCUUAGACAA-------UUCCAAAAAAUU ..(((((.(....((((((((...(((((......)))))....-(((.........)))..............)))))))))))))).............-------............ ( -18.40) >DroYak_CAF1 12269 118 + 1 AAUUGCUUGAAAAUUAUUUGCACCCCC-AACCCGAUAGAGAAAA-AGCCAAACAGAAGCAAAACCAAAAUAAACGCAAAUGACAGCAAAUCUUUUGGACAAAGAGAGGUUACACCAAAUG ..(((((.(....((((((((......-....(......)....-.((.........))...............)))))))))))))).((((((.....))))))((.....))..... ( -14.70) >consensus AAUUGCUUGAAAAUUAUUUGCACCCCCUAACCGGAUAGGGAAAA_AGCCAAAUAGAAGCAAAACCAAAAUAUACGCAAAUGACAGCAAAUCUCUUAGACAA_______UUCCAAAAAAUG ..(((((.(....((((((((...(((((......)))))......((.........))...............))))))))))))))................................ (-12.84 = -13.40 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:55 2006