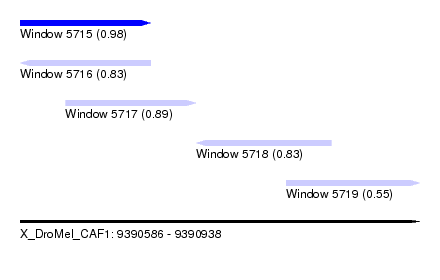
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,390,586 – 9,390,938 |
| Length | 352 |
| Max. P | 0.981613 |

| Location | 9,390,586 – 9,390,701 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.74 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -30.59 |
| Energy contribution | -31.02 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9390586 115 + 22224390 CAAUUACCAAAGUCAAUGUCAAAGUUAGCAAUCUGCCAAGUUGACGACUUAAUUUCAUGUCGAAUUGCGGAACUGAUUGCGAUUCGAUGUGACAGCGGUGAAAAGUGG-GUAAA----GU ...(((((.(((((...(((((..((.((.....)).)).))))))))))....((((((((((((((((......))))))))))))))))....))))).......-.....----.. ( -34.20) >DroSec_CAF1 4610 114 + 1 CAAUUACCACAGUCAAUGUCA-AGUUAGCAAUCUGCCAAGUUGACGACUUAAUUUCAGGUCGAAUUGCGGAACUGAUUGCGAUUCGAUGUGACAGCGGUGAAAAGUGG-GUAAA----GU ......((((((((...((((-(.((.((.....)).)).))))))))).....(((.((((((((((((......)))))))))))).)))............))))-.....----.. ( -35.60) >DroSim_CAF1 8950 114 + 1 CAAUUACCAAAGUCAAUGUCA-AGUUAGCAAUCUGCCAAGUUGACGACUUAAUUUCAUGUCGAAUUGCGGAACUCAUUGCGAUUCGACGUGACAGCGGUGAAAAGUGG-GUAAA----GU ...(((((.(((((...((((-(.((.((.....)).)).))))))))))....((((((((((((((((......))))))))))))))))....))))).......-.....----.. ( -39.40) >DroYak_CAF1 8310 119 + 1 CAAUUACCAAAGUCAAAAUCA-AGUUAACAAUCUGUCAAGUUGACGACUUAAUUUCAUGUCGUAUUGCGGACCUGAUUGCGAUUCGAUGUGACAGCGGUGAAAAGUGGUGUAGAAGUGGU .....((((...((...((((-..((.((...((((((.(((((((((..........)))))(((((((......))))))).)))).))))))..))..))..))))...))..)))) ( -29.20) >consensus CAAUUACCAAAGUCAAUGUCA_AGUUAGCAAUCUGCCAAGUUGACGACUUAAUUUCAUGUCGAAUUGCGGAACUGAUUGCGAUUCGAUGUGACAGCGGUGAAAAGUGG_GUAAA____GU ...(((((.(((((...((((.(.((.((.....)).)).))))))))))....((((((((((((((((......))))))))))))))))....)))))................... (-30.59 = -31.02 + 0.44)

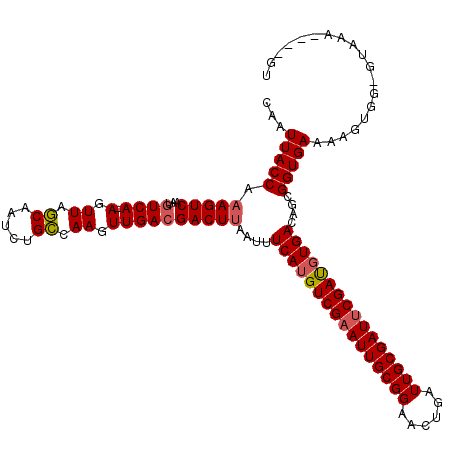

| Location | 9,390,586 – 9,390,701 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.74 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -22.69 |
| Energy contribution | -22.38 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9390586 115 - 22224390 AC----UUUAC-CCACUUUUCACCGCUGUCACAUCGAAUCGCAAUCAGUUCCGCAAUUCGACAUGAAAUUAAGUCGUCAACUUGGCAGAUUGCUAACUUUGACAUUGACUUUGGUAAUUG ..----.....-.........((((..((((..((((((.((..........)).))))))..............(((((.(((((.....)))))..)))))..))))..))))..... ( -25.10) >DroSec_CAF1 4610 114 - 1 AC----UUUAC-CCACUUUUCACCGCUGUCACAUCGAAUCGCAAUCAGUUCCGCAAUUCGACCUGAAAUUAAGUCGUCAACUUGGCAGAUUGCUAACU-UGACAUUGACUGUGGUAAUUG ..----.....-.........(((((.((((..((((((.((..........)).))))))..............(((((.(((((.....))))).)-))))..)))).)))))..... ( -30.10) >DroSim_CAF1 8950 114 - 1 AC----UUUAC-CCACUUUUCACCGCUGUCACGUCGAAUCGCAAUGAGUUCCGCAAUUCGACAUGAAAUUAAGUCGUCAACUUGGCAGAUUGCUAACU-UGACAUUGACUUUGGUAAUUG ..----.....-.........((((..((((.(((((((.((..........)).))))))).............(((((.(((((.....))))).)-))))..))))..))))..... ( -31.00) >DroYak_CAF1 8310 119 - 1 ACCACUUCUACACCACUUUUCACCGCUGUCACAUCGAAUCGCAAUCAGGUCCGCAAUACGACAUGAAAUUAAGUCGUCAACUUGACAGAUUGUUAACU-UGAUUUUGACUUUGGUAAUUG .....................((((..((((.(((((...((((((..(((......(((((..........)))))......))).))))))....)-))))..))))..))))..... ( -24.40) >consensus AC____UUUAC_CCACUUUUCACCGCUGUCACAUCGAAUCGCAAUCAGUUCCGCAAUUCGACAUGAAAUUAAGUCGUCAACUUGGCAGAUUGCUAACU_UGACAUUGACUUUGGUAAUUG .....................((((..((((..((((((.((..........)).))))))..............((((..(((((.....)))))...))))..))))..))))..... (-22.69 = -22.38 + -0.31)

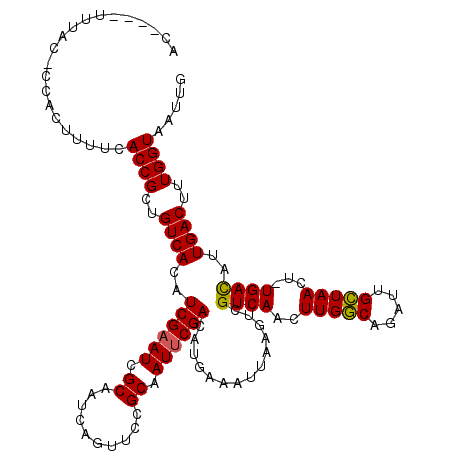

| Location | 9,390,626 – 9,390,741 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.77 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -30.37 |
| Energy contribution | -31.43 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9390626 115 + 22224390 UUGACGACUUAAUUUCAUGUCGAAUUGCGGAACUGAUUGCGAUUCGAUGUGACAGCGGUGAAAAGUGG-GUAAA----GUGUGGAAACUCUGGCAAAUACAAGCACAAGCCGCAUUCGAU ....(((.......((((((((((((((((......))))))))))))))))..(((((.....((..-(((..----.((((....)....)))..)))..))....)))))..))).. ( -35.50) >DroSec_CAF1 4649 115 + 1 UUGACGACUUAAUUUCAGGUCGAAUUGCGGAACUGAUUGCGAUUCGAUGUGACAGCGGUGAAAAGUGG-GUAAA----GUGUGGAAACUCUGGCAAAUACAAGCACAAGCCGCAUUCGAU ....(((.......(((.((((((((((((......)))))))))))).)))..(((((.....((..-(((..----.((((....)....)))..)))..))....)))))..))).. ( -34.80) >DroSim_CAF1 8989 115 + 1 UUGACGACUUAAUUUCAUGUCGAAUUGCGGAACUCAUUGCGAUUCGACGUGACAGCGGUGAAAAGUGG-GUAAA----GUGUGGAAACUCUGGCAAAUACAAGCACAAGCCGCAUUCGAU ....(((.......((((((((((((((((......))))))))))))))))..(((((.....((..-(((..----.((((....)....)))..)))..))....)))))..))).. ( -38.00) >DroYak_CAF1 8349 114 + 1 UUGACGACUUAAUUUCAUGUCGUAUUGCGGACCUGAUUGCGAUUCGAUGUGACAGCGGUGAAAAGUGGUGUAGAAGUGGUGUGGAAACUCUGGCAAAUACA------AGCCGCAUUCGAU ....(((.......((((((((.(((((((......))))))).))))))))..(((((.........((((...((.(.(.(....).)).))...))))------.)))))..))).. ( -30.50) >consensus UUGACGACUUAAUUUCAUGUCGAAUUGCGGAACUGAUUGCGAUUCGAUGUGACAGCGGUGAAAAGUGG_GUAAA____GUGUGGAAACUCUGGCAAAUACAAGCACAAGCCGCAUUCGAU ....(((.......((((((((((((((((......))))))))))))))))..(((((.....(((..(((........(.(....).).......)))...)))..)))))..))).. (-30.37 = -31.43 + 1.06)

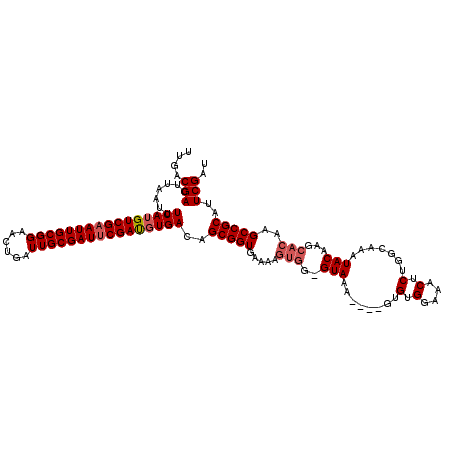
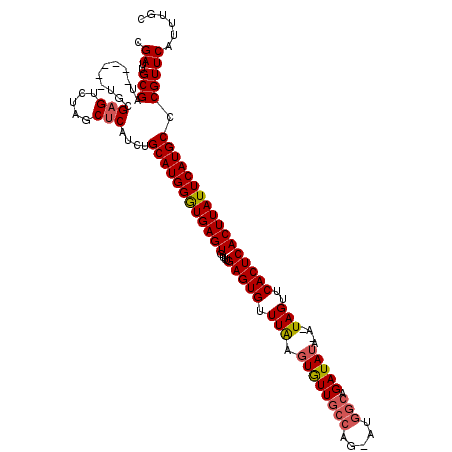
| Location | 9,390,741 – 9,390,860 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.00 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -24.96 |
| Energy contribution | -25.97 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9390741 119 - 22224390 CGAUUGCGAUUGCUAUUGCGAGUCUAGCACAUUUGCAUGGAUGAGUGUUUGAGUGUUUGAGUAUUGCCAGAAUGGCAGAUAUACA-UAGUUCACUCACUUAUUCAUGCCCGUUCAUUUGC .(((((((((....))))).))))..........(((((((((((((..(((((...((.((((((((.....)))).)))).))-..)))))..)))))))))))))............ ( -36.70) >DroSec_CAF1 4764 100 - 1 CGAUUGCGAU------UGCGAGUCUAGCUCAUCUGCAUGGGUGAGUGUUUGAGUGUUUAAGUGUUGCC---------GAUA-----UAGUUCACUCACUUAUUCAUGCCCGUUCAUUUGC .....((((.------.(((((.....)))....(((((((((((((..(((((((....(.....).---------...)-----)).))))..)))))))))))))..))....)))) ( -26.30) >DroSim_CAF1 9104 110 - 1 CGAUUGCGAU------UGCGAGUCUAGCUCAUCUGCAUGGGUGAGU----GAGUGUUUAAGUGUUGCCAGGAUGGCAGAUAUAUAGUAGUUCACUCACUUAUUCAUGCCCGUUCAUUUGC .....((((.------.(((((.....)))....((((((((((((----(((((.(((.((((((((.....)))).)))).))).....)))))))))))))))))..))....)))) ( -39.30) >consensus CGAUUGCGAU______UGCGAGUCUAGCUCAUCUGCAUGGGUGAGUGUUUGAGUGUUUAAGUGUUGCCAG_AUGGCAGAUAUA_A_UAGUUCACUCACUUAUUCAUGCCCGUUCAUUUGC .((..(((...........(((.....)))....((((((((((((....(((((.(((.((((((((.....))).)))))....)))..))))))))))))))))).)))))...... (-24.96 = -25.97 + 1.01)


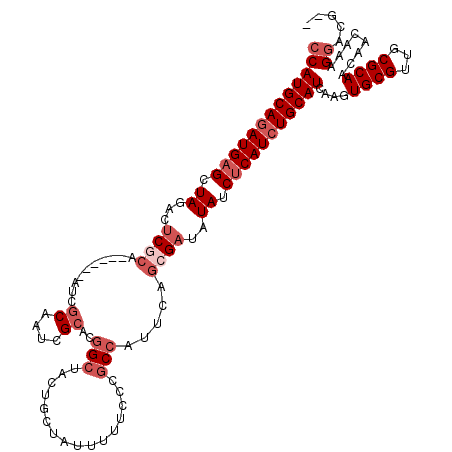
| Location | 9,390,820 – 9,390,938 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -17.85 |
| Energy contribution | -20.41 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9390820 118 + 22224390 CCAUGCAAAUGUGCUAGACUCGCAAUAGCAAUCGCAAUCGCACGGCUACUGCUAUUUUUCCCGCCAUUCAGGGAUAUAUCUCAUCUGCAUCAAGUGCGUUGCGCAACAACAAAGGACA-- ((..(((..(((((.(((...(((.((((....((....))...)))).)))......((((........)))).........)))))).))..)))((((.....))))...))...-- ( -30.30) >DroSec_CAF1 4830 112 + 1 CCAUGCAGAUGAGCUAGACUCGCA------AUCGCAAUCGCAUGGCUACUGAUAUUUUUCCCGCCAUUCAGCGAUAUAUCUCAUCUGCAUCAAGUGCGUUGCGCAACAACAAAGGACG-- (((((((((((((.((...((((.------...((....))(((((....((......))..)))))...))))..)).)))))))))))...((((...)))).........))...-- ( -35.30) >DroSim_CAF1 9180 112 + 1 CCAUGCAGAUGAGCUAGACUCGCA------AUCGCAAUCGCAUGGCUACUGCUAUUUUUCCCGCCAUUCAGCGAUAUAUCUCAUCUGCAUCAAGUGCGUUGCGCAACAACAAAGGACG-- (((((((((((((.((...((((.------...((....))(((((................)))))...))))..)).)))))))))))...((((...)))).........))...-- ( -34.59) >DroYak_CAF1 8528 102 + 1 CCAUGCAGAUGAGCUAGACCCA------------------CACAGAUACUGCCAUUUUUCCCGCCAUUCACGGAUAUAUCUCAUCUGCAUCAAGUGCGUUACGCAACAACAAAGUACGGC (((((((((((((.((......------------------..(((...))).........(((.......)))...)).)))))))))))...((((................)))))). ( -22.59) >consensus CCAUGCAGAUGAGCUAGACUCGCA______AUCGCAAUCGCACGGCUACUGCUAUUUUUCCCGCCAUUCAGCGAUAUAUCUCAUCUGCAUCAAGUGCGUUGCGCAACAACAAAGGACG__ (((((((((((((.((...((((..........((....))..(((................))).....))))..)).)))))))))))....((((...))))........))..... (-17.85 = -20.41 + 2.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:47 2006