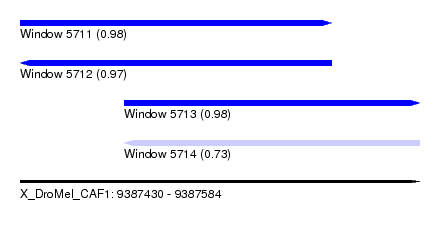
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,387,430 – 9,387,584 |
| Length | 154 |
| Max. P | 0.982173 |

| Location | 9,387,430 – 9,387,550 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -40.97 |
| Consensus MFE | -26.50 |
| Energy contribution | -26.07 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9387430 120 + 22224390 GGUAAAUAAUACGGCAGAUAUCCGCUAGCCUUGAUAUGUUCAAUGUCCACGAGGCGUAUGCUUGAUGUGUAGUAACUUUGAGCAAUAGUAGCGAGCAUCAAGCAUACGCCGCAUUGGCCC (((...((((.((((............((((((((((.....)))))...)))))(((((((((((((...((.(((.........))).))..))))))))))))))))).))))))). ( -44.80) >DroSim_CAF1 4511 118 + 1 AGUAGCUAAUACG-CUGAUAUCCGCUAACCUUGAUAUGUU-AAUGUCCACGAGGCGUAUGCUUGAUAUGUAGUAACUUCAAGUAGUAGUAGCGAGCAUCAAGCAUACGCCGCAUUGGCCC ..((((......)-)))......(((((....(((((...-.)))))...(.(((((((((((((..(((.((.(((.(.....).))).))..)))))))))))))))).).))))).. ( -37.40) >DroYak_CAF1 4907 120 + 1 GUUAAGUAAUACUGCAGAUACCCGCUACUCCUGAUACAUUUAAUGUACACGCGGCGUAUGCUUGAUAUGUGGUACAUACAAGUACUAGCAGUAAGCAUCAAGUAUACGCCGCAUUGGCCC .....(((....)))........((((....((.(((((...))))))).((((((((((((((((...((((((......))))))((.....))))))))))))))))))..)))).. ( -40.70) >consensus GGUAAAUAAUACGGCAGAUAUCCGCUAACCUUGAUAUGUU_AAUGUCCACGAGGCGUAUGCUUGAUAUGUAGUAACUUCAAGUAAUAGUAGCGAGCAUCAAGCAUACGCCGCAUUGGCCC .......................((((.....(((((.....))))).....((((((((((((((((...(......)..)))...((.....)).)))))))))))))....)))).. (-26.50 = -26.07 + -0.44)



| Location | 9,387,430 – 9,387,550 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -26.90 |
| Energy contribution | -26.36 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9387430 120 - 22224390 GGGCCAAUGCGGCGUAUGCUUGAUGCUCGCUACUAUUGCUCAAAGUUACUACACAUCAAGCAUACGCCUCGUGGACAUUGAACAUAUCAAGGCUAGCGGAUAUCUGCCGUAUUAUUUACC .((((.(((.(((((((((((((((...((.......)).....((....)).))))))))))))))).))).....((((.....)))))))).((((....)))).(((.....))). ( -41.70) >DroSim_CAF1 4511 118 - 1 GGGCCAAUGCGGCGUAUGCUUGAUGCUCGCUACUACUACUUGAAGUUACUACAUAUCAAGCAUACGCCUCGUGGACAUU-AACAUAUCAAGGUUAGCGGAUAUCAG-CGUAUUAGCUACU .(((.((((((((((((((((((((......(((.(.....).))).......)))))))))))))))((((.(((...-...........))).)))).......-.))))).)))... ( -36.96) >DroYak_CAF1 4907 120 - 1 GGGCCAAUGCGGCGUAUACUUGAUGCUUACUGCUAGUACUUGUAUGUACCACAUAUCAAGCAUACGCCGCGUGUACAUUAAAUGUAUCAGGAGUAGCGGGUAUCUGCAGUAUUACUUAAC ...((.(((((((((((.((((((((.....))..((((......)))).....)))))).)))))))))))((((((...))))))..))....((((....))))............. ( -38.40) >consensus GGGCCAAUGCGGCGUAUGCUUGAUGCUCGCUACUACUACUUGAAGUUACUACAUAUCAAGCAUACGCCUCGUGGACAUU_AACAUAUCAAGGCUAGCGGAUAUCUGCCGUAUUACUUACC .....((((((((((((((((((((.........(((......))).......)))))))))))))))...............(((((..........))))).....)))))....... (-26.90 = -26.36 + -0.55)



| Location | 9,387,470 – 9,387,584 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.90 |
| Mean single sequence MFE | -39.87 |
| Consensus MFE | -29.41 |
| Energy contribution | -28.53 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9387470 114 + 22224390 CAAUGUCCACGAGGCGUAUGCUUGAUGUGUAGUAACUUUGAGCAAUAGUAGCGAGCAUCAAGCAUACGCCGCAUUGGCCCGCCGAGACUUAAAACAUUGUGC--AAUUUCCCCC----GC ((((((......((((((((((((((((...((.(((.........))).))..))))))))))))))))(..((((....))))..).....))))))...--..........----.. ( -41.00) >DroSim_CAF1 4550 113 + 1 -AAUGUCCACGAGGCGUAUGCUUGAUAUGUAGUAACUUCAAGUAGUAGUAGCGAGCAUCAAGCAUACGCCGCAUUGGCCCGCCGAGACUUAAAACAUUGUGC--AAUUUCCCCC----AC -......((((((((((((((((((..(((.((.(((.(.....).))).))..))))))))))))))))(..((((....))))..)........))))).--..........----.. ( -34.90) >DroYak_CAF1 4947 119 + 1 UAAUGUACACGCGGCGUAUGCUUGAUAUGUGGUACAUACAAGUACUAGCAGUAAGCAUCAAGUAUACGCCGCAUUGGCCCGCCGAGAGUUA-AACAUUGUGCCCCACUUCCCCCCUGCCC ....(((((.((((((((((((((((...((((((......))))))((.....)))))))))))))))))).((((....))))......-.....))))).................. ( -43.70) >consensus _AAUGUCCACGAGGCGUAUGCUUGAUAUGUAGUAACUUCAAGUAAUAGUAGCGAGCAUCAAGCAUACGCCGCAUUGGCCCGCCGAGACUUAAAACAUUGUGC__AAUUUCCCCC____AC ((((((......((((((((((((((((...(......)..)))...((.....)).)))))))))))))...((((....))))........))))))..................... (-29.41 = -28.53 + -0.88)

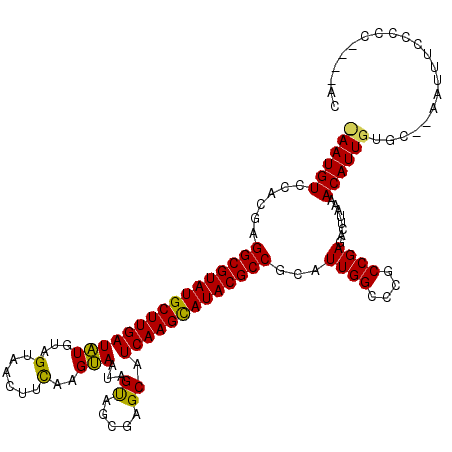
| Location | 9,387,470 – 9,387,584 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.90 |
| Mean single sequence MFE | -39.87 |
| Consensus MFE | -29.63 |
| Energy contribution | -29.52 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9387470 114 - 22224390 GC----GGGGGAAAUU--GCACAAUGUUUUAAGUCUCGGCGGGCCAAUGCGGCGUAUGCUUGAUGCUCGCUACUAUUGCUCAAAGUUACUACACAUCAAGCAUACGCCUCGUGGACAUUG ((----((......))--)).((((((((........((....)).(((.(((((((((((((((...((.......)).....((....)).))))))))))))))).))))))))))) ( -40.30) >DroSim_CAF1 4550 113 - 1 GU----GGGGGAAAUU--GCACAAUGUUUUAAGUCUCGGCGGGCCAAUGCGGCGUAUGCUUGAUGCUCGCUACUACUACUUGAAGUUACUACAUAUCAAGCAUACGCCUCGUGGACAUU- ((----.((((.....--((.....))......)))).))(..(((.((.(((((((((((((((......(((.(.....).))).......))))))))))))))).))))).)...- ( -36.72) >DroYak_CAF1 4947 119 - 1 GGGCAGGGGGGAAGUGGGGCACAAUGUU-UAACUCUCGGCGGGCCAAUGCGGCGUAUACUUGAUGCUUACUGCUAGUACUUGUAUGUACCACAUAUCAAGCAUACGCCGCGUGUACAUUA .(((.(..((((.((..(((.....)))-..))))))..)..))).(((((((((((.((((((((.....))..((((......)))).....)))))).)))))))))))........ ( -42.60) >consensus GC____GGGGGAAAUU__GCACAAUGUUUUAAGUCUCGGCGGGCCAAUGCGGCGUAUGCUUGAUGCUCGCUACUACUACUUGAAGUUACUACAUAUCAAGCAUACGCCUCGUGGACAUU_ ........((((......((.....))......))))((....)).(((.(((((((((((((((.........(((......))).......))))))))))))))).)))........ (-29.63 = -29.52 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:43 2006