| Sequence ID | X_DroMel_CAF1 |
|---|---|
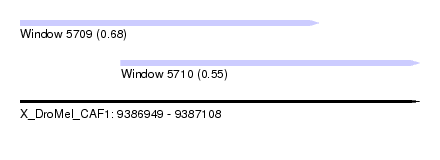
| Location | 9,386,949 – 9,387,108 |
| Length | 159 |
| Max. P | 0.680837 |

| Location | 9,386,949 – 9,387,068 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.98 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -14.57 |
| Energy contribution | -15.88 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.680837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
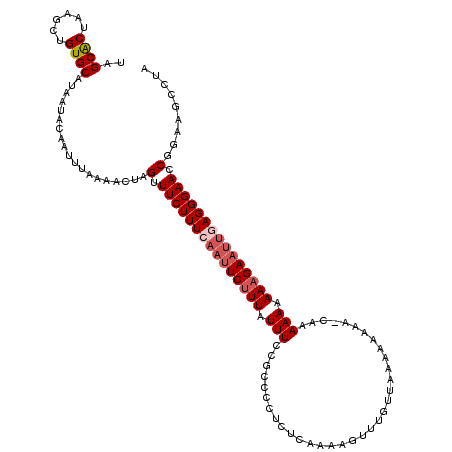
>X_DroMel_CAF1 9386949 119 + 22224390 UAGCACUAAGCUGUGCAUAAUAAAAUUUAAAACUAGUUUCUUUCAAUUCUUUAUUUCCGCCUCCCUCAAAAGUUUAACAAAAAAUA-AAAAAAAAAAGAAUUAAGGGAACCGGAAGCCUA ..((((......)))).....................................((((((..(((((....(((((...........-..........))))).)))))..)))))).... ( -18.20) >DroSec_CAF1 956 120 + 1 UAGCACUAAGCUGUGCAUAAUACAAUUUAAAACUAGUUUCUUUCAAUUCUUUAUUUUCGCCCCUAUCAAAAGUUUUUAAAAAAAAACCAAAAAAAAAGAAUUGAGGGAACCGGAAGCCUA ..((((......))))................(..(((.(((((((((((((.((((.(............((((((....))))))).)))).))))))))))))))))..)....... ( -23.40) >DroSim_CAF1 4032 119 + 1 UAGCACUAAGCUGUGCAUAAUACAAUUUAAAACUAGUUUCUUUCAAUUCUUUAUUUCCGCCCCUCUCAAAAGUUUGUUAAAAAAAA-CAAAAAAAAAGAAUUGAGGGAACCGGAAGCCUA ..((((......))))................(..(((.(((((((((((((..........((......))((((((......))-))))...))))))))))))))))..)....... ( -24.20) >DroYak_CAF1 4441 110 + 1 UAGCGCUAAGCUGUGCAUAAUACAAUUUAAAACUAGUUUCUUUCAAUUCCUUAUUUCCGCCCCUCUCAAAAGUUUGUUAACG------AAAAAAAAAGA----AGGGAAACGGAAGCCUA ..((((......)))).....................................(((((((((.(((......((((....))------))......)))----.)))...)))))).... ( -19.40) >consensus UAGCACUAAGCUGUGCAUAAUACAAUUUAAAACUAGUUUCUUUCAAUUCUUUAUUUCCGCCCCUCUCAAAAGUUUGUUAAAAAAAA_CAAAAAAAAAGAAUUGAGGGAACCGGAAGCCUA ..((((......))))...................(.(((((((((((((((.(((..................................))).))))))))))))))).)......... (-14.57 = -15.88 + 1.31)


| Location | 9,386,989 – 9,387,108 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.01 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -18.78 |
| Energy contribution | -19.97 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
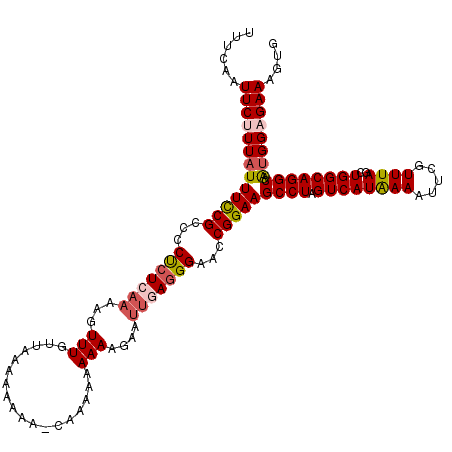
>X_DroMel_CAF1 9386989 119 + 22224390 UUUCAAUUCUUUAUUUCCGCCUCCCUCAAAAGUUUAACAAAAAAUA-AAAAAAAAAAGAAUUAAGGGAACCGGAAGCCUAGUCAUAAAAUUCGUUUAGCUGGCAGGCAAUGGUGAAAGUG (((((.........(((((..(((((....(((((...........-..........))))).)))))..)))))((((.((((((((.....))))..)))))))).....)))))... ( -23.20) >DroSec_CAF1 996 120 + 1 UUUCAAUUCUUUAUUUUCGCCCCUAUCAAAAGUUUUUAAAAAAAAACCAAAAAAAAAGAAUUGAGGGAACCGGAAGCCUAGUCAUAAAAUUCGUUUAGCUGGCAGGCAAUGGAGAAAGUG ......(((((((((((((.((((.......((((((....))))))(((..........)))))))...)))).((((.((((((((.....))))..))))))))))))))))).... ( -22.80) >DroSim_CAF1 4072 119 + 1 UUUCAAUUCUUUAUUUCCGCCCCUCUCAAAAGUUUGUUAAAAAAAA-CAAAAAAAAAGAAUUGAGGGAACCGGAAGCCUAGUCAUGAAAUUCGUUUAGCUGGCAGGCAAAGGAGAAAGUG ((((..((((((..(((((.(((((((.....((((((......))-))))......))...)))))...)))))((((.((((..(((....)))...))))))))))))))))))... ( -29.10) >DroYak_CAF1 4481 110 + 1 UUUCAAUUCCUUAUUUCCGCCCCUCUCAAAAGUUUGUUAACG------AAAAAAAAAGA----AGGGAAACGGAAGCCUAGUCAUGAAAUUCGUUUAGCUGGCAGGCAGUGGAGAAAGUG ......((((....((((((((.(((......((((....))------))......)))----.)))...)))))((((.((((..(((....)))...))))))))...))))...... ( -26.00) >consensus UUUCAAUUCUUUAUUUCCGCCCCUCUCAAAAGUUUGUUAAAAAAAA_CAAAAAAAAAGAAUUGAGGGAACCGGAAGCCUAGUCAUAAAAUUCGUUUAGCUGGCAGGCAAUGGAGAAAGUG ......(((((((((((((...(((((((...(((..................)))....)))))))...)))))((((.((((((((.....))))..)))))))).)))))))).... (-18.78 = -19.97 + 1.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:39 2006