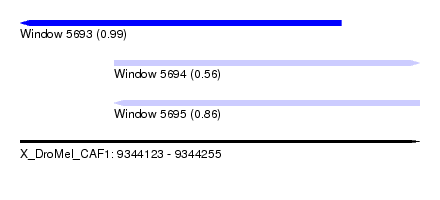
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,344,123 – 9,344,255 |
| Length | 132 |
| Max. P | 0.990331 |

| Location | 9,344,123 – 9,344,229 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.59 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -14.36 |
| Energy contribution | -14.76 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9344123 106 - 22224390 UUCGAGUGCAAAUUUGACUGGCU--UGGAAAAGUCCAGGCGAGCAAAUUGGCCAGAGGUGUUGCCAAC-GAAAAAAAA---AAAAAAAAAUGAAUUGGUAACGAAAACAAUU (((..(.((.((((((.((.(((--((((....))))))).)))))))).)))......((((((((.-.........---.............)))))))))))....... ( -29.20) >DroSec_CAF1 124247 101 - 1 UUCGAGUGCAAAUUUGGCUGGCU--UGGAAAAGUCCAGGCCAGCAAAUUGGCCAGAGGUGUUGCCAAC-GAAAAAAAA---AAGAAAAA-----UUGAUAAAGAAAACAAUU ((((.(.((.((((((.((((((--((((....)))))))))))))))).)))...((.....))..)-)))......---........-----.................. ( -30.10) >DroSim_CAF1 121639 100 - 1 UUCGAGUGCAAAUUUGGCUGGCU--UGCAAAAGUCCAGGCGAGCAAAUUGGCCAGAGGUGUUGCCAAC-AAAAAAAAU---AAAAAAAA------UGGUAAAGAUAACAAUU .......(((..(((((((.(((--(((..........)))))).....)))))))..)))(((((..-.........---........------)))))............ ( -22.31) >DroEre_CAF1 133230 93 - 1 UUCUAGUGCACAUUUGACUGGCUUUUGGAAAAGUCCAGGCGAGCAAAUUGGCCAGAGGUAUUGCAAGC--------------GAAAAAA-----UUGCCAAAGAAAACAAUU ((((..((((((((((.((.(((..((((....))))))).)))))).))(((...)))..)))).((--------------((.....-----))))...))))....... ( -19.80) >DroYak_CAF1 127146 105 - 1 UUCGAGUGCAAAUUUGACUGGCU--UGGAAAAGUCCAGGCGAGCAAAUUGGCCAGUGCAAAUGCAAUACAAAAUGAAUUUCAAAAAAAA-----UUGGCAACGAAAACAAUU ((((......((((((.((.(((--((((....))))))).)))))))).((((((((....)).........((.....))......)-----)))))..))))....... ( -29.60) >consensus UUCGAGUGCAAAUUUGACUGGCU__UGGAAAAGUCCAGGCGAGCAAAUUGGCCAGAGGUGUUGCCAAC_AAAAAAAAA___AAAAAAAA_____UUGGUAAAGAAAACAAUU .....(.((.((((((.((.(((..((((....))))))).)))))))).)))........................................................... (-14.36 = -14.76 + 0.40)

| Location | 9,344,154 – 9,344,255 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.94 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9344154 101 + 22224390 --UUUUUUUUC-GUUGGCAACACCUCUGGCCAAUUUGCUCGCCUGGACUUUUCCA--AGCCAGUCAAAUUUGCACUCGAAGUGGAAAACUUUCGACUGU-----------GAGUGUU --.........-((((((..........))))))(((((.((.((((....))))--.)).)).)))....(((((((.(((.(((....))).))).)-----------)))))). ( -28.80) >DroSec_CAF1 124273 101 + 1 --UUUUUUUUC-GUUGGCAACACCUCUGGCCAAUUUGCUGGCCUGGACUUUUCCA--AGCCAGCCAAAUUUGCACUCGAAGUGGAAAACUUUCGACUGU-----------GAGUGUU --.........-((((((..........))))))..((((((.((((....))))--.)))))).......(((((((.(((.(((....))).))).)-----------)))))). ( -36.40) >DroSim_CAF1 121664 101 + 1 --AUUUUUUUU-GUUGGCAACACCUCUGGCCAAUUUGCUCGCCUGGACUUUUGCA--AGCCAGCCAAAUUUGCACUCGAAGUGGAAAACUUUCGACUGU-----------GAGUGUU --.........-.(((((.........(((..........)))(((.(((....)--))))))))))....(((((((.(((.(((....))).))).)-----------)))))). ( -26.80) >DroEre_CAF1 133255 94 + 1 ------------GCUUGCAAUACCUCUGGCCAAUUUGCUCGCCUGGACUUUUCCAAAAGCCAGUCAAAUGUGCACUAGAAGUGGAAAACUUUCGACUGC-----------GAGUGUU ------------((((((......(((((...(((((((.((.((((....))))...)).)).))))).....)))))(((.(((....))).)))))-----------))))... ( -24.90) >DroYak_CAF1 127173 115 + 1 AAAUUCAUUUUGUAUUGCAUUUGCACUGGCCAAUUUGCUCGCCUGGACUUUUCCA--AGCCAGUCAAAUUUGCACUCGAAGUGGAAAACUUUCGACUGUGUGUGUGUGUUGAGUGUU ...(((((((((...((((((((.((((((......(....).((((....))))--.))))))))))..))))..))))))))).(((.((((((...........)))))).))) ( -30.10) >consensus __AUUUUUUUC_GUUGGCAACACCUCUGGCCAAUUUGCUCGCCUGGACUUUUCCA__AGCCAGUCAAAUUUGCACUCGAAGUGGAAAACUUUCGACUGU___________GAGUGUU ............((((((.........(((..........)))((((....))))...)))))).......((((((..(((.(((....))).))).............)))))). (-17.02 = -17.94 + 0.92)


| Location | 9,344,154 – 9,344,255 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -18.02 |
| Energy contribution | -19.62 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9344154 101 - 22224390 AACACUC-----------ACAGUCGAAAGUUUUCCACUUCGAGUGCAAAUUUGACUGGCU--UGGAAAAGUCCAGGCGAGCAAAUUGGCCAGAGGUGUUGCCAAC-GAAAAAAAA-- .......-----------....(((...((....((((((..(.((.((((((.((.(((--((((....))))))).)))))))).))).))))))..))...)-)).......-- ( -30.20) >DroSec_CAF1 124273 101 - 1 AACACUC-----------ACAGUCGAAAGUUUUCCACUUCGAGUGCAAAUUUGGCUGGCU--UGGAAAAGUCCAGGCCAGCAAAUUGGCCAGAGGUGUUGCCAAC-GAAAAAAAA-- ..(((((-----------..(((.(((....))).)))..)))))....(((((((((((--((((....)))))))))))...(((((..........))))))-)))......-- ( -35.40) >DroSim_CAF1 121664 101 - 1 AACACUC-----------ACAGUCGAAAGUUUUCCACUUCGAGUGCAAAUUUGGCUGGCU--UGCAAAAGUCCAGGCGAGCAAAUUGGCCAGAGGUGUUGCCAAC-AAAAAAAAU-- ((((((.-----------.((.(((((.((.....))))))).))....(((((((.(((--(((..........)))))).....)))))))))))))......-.........-- ( -27.80) >DroEre_CAF1 133255 94 - 1 AACACUC-----------GCAGUCGAAAGUUUUCCACUUCUAGUGCACAUUUGACUGGCUUUUGGAAAAGUCCAGGCGAGCAAAUUGGCCAGAGGUAUUGCAAGC------------ .......-----------((((((....)......((((((.(.((..(((((.((.(((..((((....))))))).)))))))..))))))))))))))....------------ ( -26.00) >DroYak_CAF1 127173 115 - 1 AACACUCAACACACACACACAGUCGAAAGUUUUCCACUUCGAGUGCAAAUUUGACUGGCU--UGGAAAAGUCCAGGCGAGCAAAUUGGCCAGUGCAAAUGCAAUACAAAAUGAAUUU ....(((............((((((((...(((.(((.....))).)))))))))))(((--((((....))))))))))....(((.....(((....)))...)))......... ( -27.70) >consensus AACACUC___________ACAGUCGAAAGUUUUCCACUUCGAGUGCAAAUUUGACUGGCU__UGGAAAAGUCCAGGCGAGCAAAUUGGCCAGAGGUGUUGCCAAC_AAAAAAAAA__ ((((((................(((((.((.....)))))))(.((.((((((.((.(((..((((....))))))).)))))))).)))...)))))).................. (-18.02 = -19.62 + 1.60)

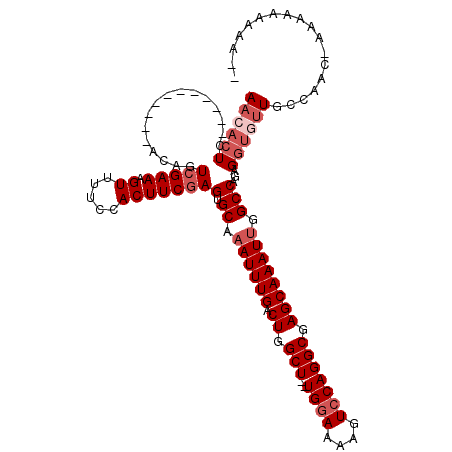
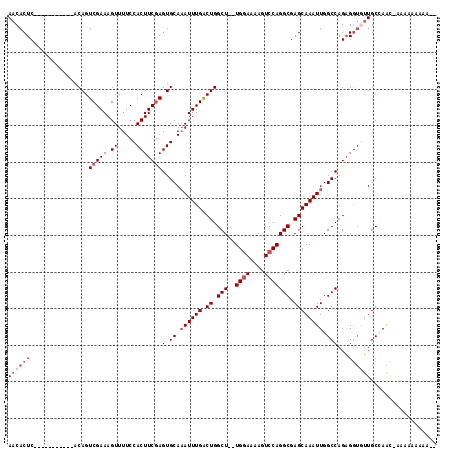
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:25 2006