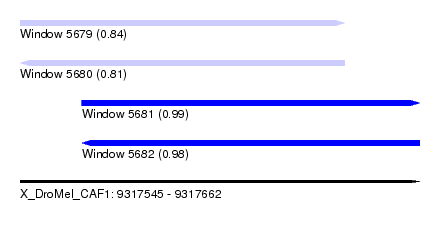
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,317,545 – 9,317,662 |
| Length | 117 |
| Max. P | 0.993762 |

| Location | 9,317,545 – 9,317,640 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -41.55 |
| Consensus MFE | -26.61 |
| Energy contribution | -27.17 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9317545 95 + 22224390 -------GCUGAUGGUGGUCGCCAGCGAGAACGCAUUCUGGCCUUGCUGG---GCGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUU -------...(((..(((((((.(((.((((....)))).((((....))---)).))).))))))).))).((((((.(((((....))))).))))))..... ( -43.70) >DroVir_CAF1 118873 95 + 1 -------GCUGUUGGUGGCGGCCAUCGGGAACGCAUCUUGGCAUUGUUGG---GUGGUUUGCGGCCGCAUCCGAAUCCAAUGCCCACGGGCCUCGGAUUCAUUUU -------......((..((((((...(.((((.((.((..((...))..)---))))))).)))))))..))((((((...(((....)))...))))))..... ( -38.00) >DroPse_CAF1 110185 95 + 1 ----------GGUCGGGGUCGCCAGCGGGAGCGCAUUUUUGCGUUGUUGGGGGGCGGCUUGCGACCAAACCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUC ----------(((((((((((((..(((.((((((....)))))).)))...)))))))).)))))......((((((.(((((....))))).))))))..... ( -50.80) >DroGri_CAF1 114225 102 + 1 UAGUGGUGCAGGUGGUGGCGGCCAUCGGGAACGCAUCUUGGCAUUGUUGG---GUGGUUUGCGGCCGCAUCCGAAUCCAAUGCCCACGGGCCUCGGAUUCAUUUU ....((.....(((((.((((((((((....)((......)).......)---)))).)))).)))))..))((((((...(((....)))...))))))..... ( -39.00) >DroWil_CAF1 110866 95 + 1 -------GCGACUGGUGGCGGCCAACGUGAGCGCAUUUUGGCCUUAUUAG---GUGGCUUACGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUU -------((.((((((((.((((((.(((....))).)))))))))))).---)).))..............((((((.(((((....))))).))))))..... ( -37.10) >DroMoj_CAF1 126011 95 + 1 -------AGUGUUGGUGGGGGCCACCGGGAGCGCUUCUUGGCAUUGUUGG---GAGGUUUGCGGCCGCAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUU -------.((((.(((((...)))))((..((((((((..((...))..)---)))))..))..))))))..((((((.(((((....))))).))))))..... ( -40.70) >consensus _______GCUGGUGGUGGCGGCCAGCGGGAACGCAUCUUGGCAUUGUUGG___GUGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUU ............((((.((((((...(....)((......)).............))).))).)))).....((((((.(((((....))))).))))))..... (-26.61 = -27.17 + 0.56)

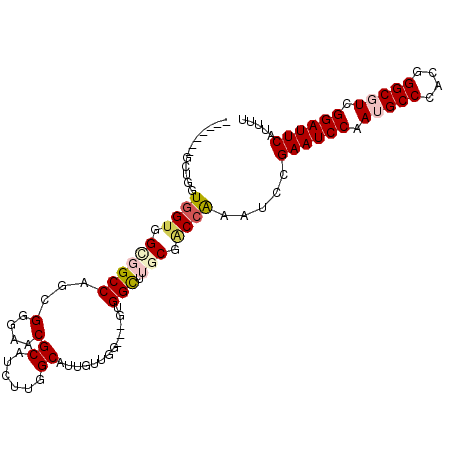

| Location | 9,317,545 – 9,317,640 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -22.98 |
| Energy contribution | -23.07 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9317545 95 - 22224390 AAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCGC---CCAGCAAGGCCAGAAUGCGUUCUCGCUGGCGACCACCAUCAGC------- ....(((((((((.(((....))).)))))))))(((.(((((((.(((.((---(......))).((((....)))).))).)))))))..)))...------- ( -39.60) >DroVir_CAF1 118873 95 - 1 AAAAUGAAUCCGAGGCCCGUGGGCAUUGGAUUCGGAUGCGGCCGCAAACCAC---CCAACAAUGCCAAGAUGCGUUCCCGAUGGCCGCCACCAACAGC------- .....((((((((.(((....))).))))))))((..(((((((.....)..---.....(((((......)))))......))))))..))......------- ( -33.10) >DroPse_CAF1 110185 95 - 1 GAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGGUUUGGUCGCAAGCCGCCCCCCAACAACGCAAAAAUGCGCUCCCGCUGGCGACCCCGACC---------- ....(((((((((.(((....))).)))))))))((((.((((((.(((.(........)..((((....)))).....))).))))))..))))---------- ( -37.10) >DroGri_CAF1 114225 102 - 1 AAAAUGAAUCCGAGGCCCGUGGGCAUUGGAUUCGGAUGCGGCCGCAAACCAC---CCAACAAUGCCAAGAUGCGUUCCCGAUGGCCGCCACCACCUGCACCACUA .....((((((((.(((....))).))))))))((..(((((((.....)..---.....(((((......)))))......))))))..))............. ( -33.10) >DroWil_CAF1 110866 95 - 1 AAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGUAAGCCAC---CUAAUAAGGCCAAAAUGCGCUCACGUUGGCCGCCACCAGUCGC------- .....((((((((.(((....))).))))))))((...(((.(....)))).---.......((((((..((....))..))))))....))......------- ( -34.10) >DroMoj_CAF1 126011 95 - 1 AAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUGCGGCCGCAAACCUC---CCAACAAUGCCAAGAAGCGCUCCCGGUGGCCCCCACCAACACU------- .....((((((((.(((....))).))))))))(((.((((.......))..---........((......))))))).(((((...)))))......------- ( -31.20) >consensus AAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUGCGGCCGCAAACCAC___CCAACAAUGCCAAAAUGCGCUCCCGAUGGCCGCCACCAACAGC_______ .....((((((((.(((....))).))))))))((...(((((....................((......))(....)...)))))...))............. (-22.98 = -23.07 + 0.08)

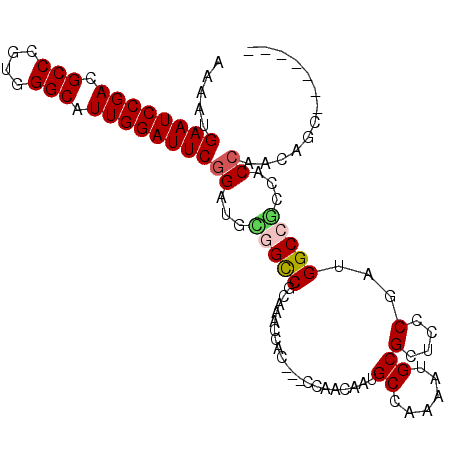

| Location | 9,317,563 – 9,317,662 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.51 |
| Mean single sequence MFE | -42.22 |
| Consensus MFE | -21.90 |
| Energy contribution | -22.32 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9317563 99 + 22224390 CGAGAACGCAUUCUGGCCUUGCUGG---GCGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUCG---UCGGGCG-CU-UCGCUU------------- ...(((.((......((((....))---)).(((((((((((((...((((((.(((((....))))).))))))...))))).)))---.))))))-))-))....------------- ( -42.90) >DroPse_CAF1 110200 102 + 1 CGGGAGCGCAUUUUUGCGUUGUUGGGGGGCGGCUUGCGACCAAACCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUC-GGUCGG---AAUGUCG-UUCUCGCUC------------- (((.((((((....)))))).)))(((((((((((.(((((......((((((.(((((....))))).)))))).....-))))).---)).))))-)))))....------------- ( -49.50) >DroWil_CAF1 110884 102 + 1 CGUGAGCGCAUUUUGGCCUUAUUAG---GUGGCUUACGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUGGUUUCCACUUUAGACC-CU-CUUCUU------------- (((((((.(.....)((((....))---)).)))))))((((((...((((((.(((((....))))).))))))..))))))..............-..-......------------- ( -34.90) >DroYak_CAF1 98448 101 + 1 CGAGAACGCAUUCUGGCCUUGCUGG---GCGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUCGUCGUCGGGCG-CU-UCGCC-------------- (((..(((((..(((.((.....))---.)))..)))(((((((...((((((.(((((....))))).))))))...))))))).))..)))((((-..-.))))-------------- ( -43.90) >DroMoj_CAF1 126029 100 + 1 CGGGAGCGCUUCUUGGCAUUGUUGG---GAGGUUUGCGGCCGCAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUGGCGUUGUCUUUGGCCA-CU-UGUC--------------- ((((.((((((((..((...))..)---)))))..))((((......((((((.(((((....))))).)))))).....(((...)))...)))).-))-))..--------------- ( -39.40) >DroAna_CAF1 110196 112 + 1 CGGGAACGCAUUCUGGCCUUGUUGG---GCGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUGAG---UUGGUCCUCU-UUGU-AGUUUUUCUCUCUG .(((((((((..(((.((.....))---.)))..))))((((((...((((((.(((((....))))).))))))...))))))(((---......))).-....-.....))))).... ( -42.70) >consensus CGGGAACGCAUUCUGGCCUUGUUGG___GCGGCUUGCGACCAAAUCCGAAUCCAAUGCCCACGGGCGUCGGAUUCAUUUUUGGUUCG___UUGGGCG_CU_UCGCU______________ ......((((....((((..((......)))))))))).........((((((.(((((....))))).))))))............................................. (-21.90 = -22.32 + 0.42)


| Location | 9,317,563 – 9,317,662 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.51 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -21.67 |
| Energy contribution | -21.75 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9317563 99 - 22224390 -------------AAGCGA-AG-CGCCCGA---CGAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCGC---CCAGCAAGGCCAGAAUGCGUUCUCG -------------...(((-((-(((..(.---(((.(((((..(((((((((.(((....))).)))))))))..)))))))))..(((..---.......)))......))))).))) ( -38.30) >DroPse_CAF1 110200 102 - 1 -------------GAGCGAGAA-CGACAUU---CCGACC-GAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGGUUUGGUCGCAAGCCGCCCCCCAACAACGCAAAAAUGCGCUCCCG -------------((((.....-((...((---.(((((-(((.(((((((((.(((....))).)))))))))..)))))))).))..))............(((....)))))))... ( -38.60) >DroWil_CAF1 110884 102 - 1 -------------AAGAAG-AG-GGUCUAAAGUGGAAACCAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGUAAGCCAC---CUAAUAAGGCCAAAAUGCGCUCACG -------------......-..-(((((...((((..((((((.(((((((((.(((....))).)))))))))..))))))......))))---......))))).............. ( -34.20) >DroYak_CAF1 98448 101 - 1 --------------GGCGA-AG-CGCCCGACGACGAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCGC---CCAGCAAGGCCAGAAUGCGUUCUCG --------------((((.-..-))))(((.(((((.(((((..(((((((((.(((....))).)))))))))..)))))))(((..(.((---(......)))..)..)))))).))) ( -41.00) >DroMoj_CAF1 126029 100 - 1 ---------------GACA-AG-UGGCCAAAGACAACGCCAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUGCGGCCGCAAACCUC---CCAACAAUGCCAAGAAGCGCUCCCG ---------------....-.(-(((((...(....)((.....(((((((((.(((....))).)))))))))...)))))))).......---........((......))....... ( -30.90) >DroAna_CAF1 110196 112 - 1 CAGAGAGAAAAACU-ACAA-AGAGGACCAA---CUCACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCGC---CCAACAAGGCCAGAAUGCGUUCCCG ....(.(((.....-....-.(((......---)))((((((..(((((((((.(((....))).)))))))))..))))))((((..(.((---(......)))..)..))))))).). ( -36.70) >consensus ______________AGCGA_AG_CGACCAA___CGAACCAAAAAUGAAUCCGACGCCCGUGGGCAUUGGAUUCGGAUUUGGUCGCAAGCCGC___CCAACAAGGCCAAAAUGCGCUCCCG .............................................((((((((.(((....))).))))))))((...(((.(....))))....))......((......))....... (-21.67 = -21.75 + 0.08)

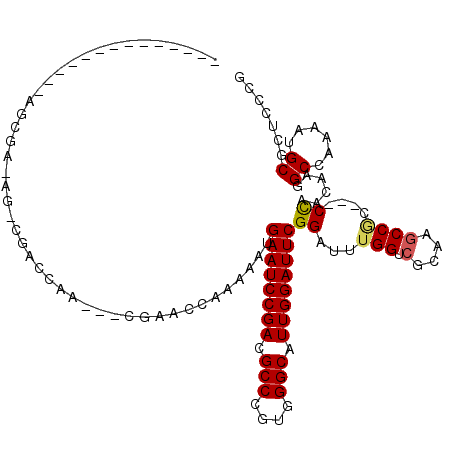
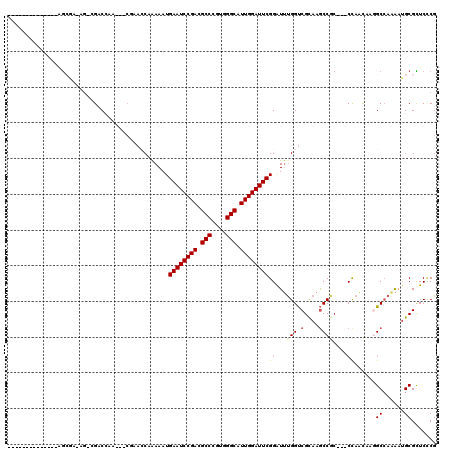
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:14 2006