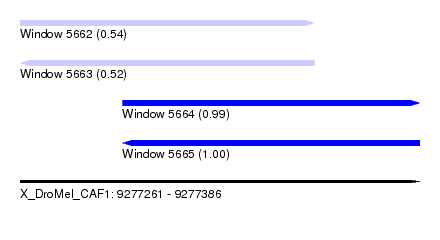
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,277,261 – 9,277,386 |
| Length | 125 |
| Max. P | 0.995131 |

| Location | 9,277,261 – 9,277,353 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -17.68 |
| Energy contribution | -18.08 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9277261 92 + 22224390 AGGAUGCUCAGGAUGUU--------UCGGGAUGCUGAGUAUGGUAAGGCAGUGUAAUUGCCGCCGGGCAGCGCUAAUGUUGCAUGUUGCUGGCGUAAUUG ...((((((((.((.(.--------...).)).)))))))).....((((((...))))))(((((.((((((.......))..)))))))))....... ( -31.60) >DroSec_CAF1 59692 91 + 1 AGG---------AUGUUCAGGAUGCUGAGCAUGCUGAGGAUGGUAAGGCAGUGUAAUUGCCGCCGGGCAGCGCUAAUGUUGCAUGUUGCUGGCGUAAUUG ((.---------((((((((....)))))))).))...........((((((...))))))(((((.((((((.......))..)))))))))....... ( -32.20) >DroSim_CAF1 57046 100 + 1 AGGAUGCUCAGGAUGUUCAGGAUGCUGAGCAUGCUGGGGAUGGUAAGGCAGUGUAAUUGCCGCCGGGCAGCGCUAAUGUUGCAUGUUGCUGGCGUAAUUG ...((.(((((.((((((((....)))))))).))))).)).....((((((...))))))(((((.((((((.......))..)))))))))....... ( -40.40) >DroEre_CAF1 66187 82 + 1 UAGAUGCUCAGGAUGUU------------------CGCGAUGGUAAGGCAGUGUAAUUGCCGCCGGGCAGCGCUAAUGUUGCAUGUUGCUGGCGUAAUUG ............((((.------------------.((((((((..((((((...)))))))))..((((((....))))))..)))))..))))..... ( -23.90) >DroYak_CAF1 58221 77 + 1 -----GCUCAGGAUGGU------------------AAGGAUGGCAAGGCAGUGUAAUUGCCACAGGGCAGCGCUAAUGUUGCAUGUUGCUGGCGUAAUUG -----((.(((......------------------...........((((((...))))))(((..((((((....)))))).)))..)))))....... ( -19.70) >consensus AGGAUGCUCAGGAUGUU________U__G_AUGCUGAGGAUGGUAAGGCAGUGUAAUUGCCGCCGGGCAGCGCUAAUGUUGCAUGUUGCUGGCGUAAUUG ..............................................((((((...))))))(((((.((((((.......))..)))))))))....... (-17.68 = -18.08 + 0.40)

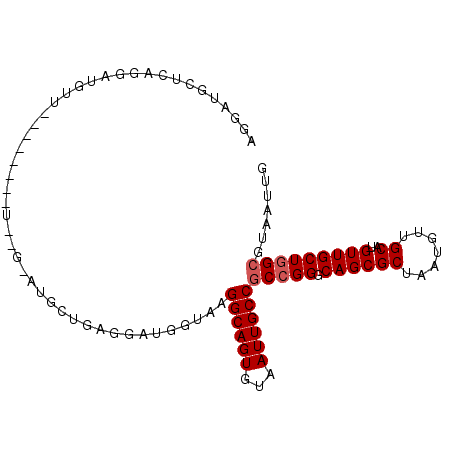

| Location | 9,277,261 – 9,277,353 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -17.98 |
| Consensus MFE | -11.70 |
| Energy contribution | -11.74 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9277261 92 - 22224390 CAAUUACGCCAGCAACAUGCAACAUUAGCGCUGCCCGGCGGCAAUUACACUGCCUUACCAUACUCAGCAUCCCGA--------AACAUCCUGAGCAUCCU .......(((.(((...(((.......))).)))..)))((((.......))))........(((((.((.....--------...)).)))))...... ( -17.80) >DroSec_CAF1 59692 91 - 1 CAAUUACGCCAGCAACAUGCAACAUUAGCGCUGCCCGGCGGCAAUUACACUGCCUUACCAUCCUCAGCAUGCUCAGCAUCCUGAACAU---------CCU .......(((.(((...(((.......))).)))..)))((((.......)))).........((((.((((...)))).))))....---------... ( -20.50) >DroSim_CAF1 57046 100 - 1 CAAUUACGCCAGCAACAUGCAACAUUAGCGCUGCCCGGCGGCAAUUACACUGCCUUACCAUCCCCAGCAUGCUCAGCAUCCUGAACAUCCUGAGCAUCCU .......(((.(((...(((.......))).)))..)))((((.......))))...........((.((((((((.((.......)).)))))))).)) ( -24.00) >DroEre_CAF1 66187 82 - 1 CAAUUACGCCAGCAACAUGCAACAUUAGCGCUGCCCGGCGGCAAUUACACUGCCUUACCAUCGCG------------------AACAUCCUGAGCAUCUA .......((((((.....)).......(((.((...(((((........)))))....)).))).------------------.......)).))..... ( -16.00) >DroYak_CAF1 58221 77 - 1 CAAUUACGCCAGCAACAUGCAACAUUAGCGCUGCCCUGUGGCAAUUACACUGCCUUGCCAUCCUU------------------ACCAUCCUGAGC----- .......(((((......(((.(......).)))...(((((((..........)))))))....------------------......))).))----- ( -11.60) >consensus CAAUUACGCCAGCAACAUGCAACAUUAGCGCUGCCCGGCGGCAAUUACACUGCCUUACCAUCCUCAGCAU_C__A________AACAUCCUGAGCAUCCU .......(((.(((...(((.......))).)))..)))((((.......)))).............................................. (-11.70 = -11.74 + 0.04)


| Location | 9,277,293 – 9,277,386 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -30.34 |
| Energy contribution | -30.98 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9277293 93 + 22224390 UGGUAAGGCAGUGUAAUUGCCGCCGGGCAGCGCUAAUGUUGCAUGUUGCUGGCGUAAUUGUUGCAGUUAAAUGCCGCCAGCAAUCAGAACCGC .(((..((((((...)))))))))((((((((....))))))..((((((((((........(((......))))))))))))).....)).. ( -34.90) >DroSec_CAF1 59723 93 + 1 UGGUAAGGCAGUGUAAUUGCCGCCGGGCAGCGCUAAUGUUGCAUGUUGCUGGCGUAAUUGUUGCAGUUAAAUGCCGCCAGCAAUCAGAACCGC .(((..((((((...)))))))))((((((((....))))))..((((((((((........(((......))))))))))))).....)).. ( -34.90) >DroSim_CAF1 57086 93 + 1 UGGUAAGGCAGUGUAAUUGCCGCCGGGCAGCGCUAAUGUUGCAUGUUGCUGGCGUAAUUGUUGCAGUUAAAUGCCGCCAGCAAUCAGAACCGC .(((..((((((...)))))))))((((((((....))))))..((((((((((........(((......))))))))))))).....)).. ( -34.90) >DroEre_CAF1 66209 93 + 1 UGGUAAGGCAGUGUAAUUGCCGCCGGGCAGCGCUAAUGUUGCAUGUUGCUGGCGUAAUUGUUGCAGCUAAAUGCCGCAAGCAAUCAGAACAGC .(((..((((((...)))))))))..((((((....)))))).....((((.....((((((((.((.....)).)).)))))).....)))) ( -28.80) >DroYak_CAF1 58238 93 + 1 UGGCAAGGCAGUGUAAUUGCCACAGGGCAGCGCUAAUGUUGCAUGUUGCUGGCGUAAUUGUUGCAGUUAAAUGCCGCCAGCGAUCAGAACCGC ..((..((((((...))))))...((((((((....))))))..((((((((((........(((......))))))))))))).....)))) ( -33.00) >consensus UGGUAAGGCAGUGUAAUUGCCGCCGGGCAGCGCUAAUGUUGCAUGUUGCUGGCGUAAUUGUUGCAGUUAAAUGCCGCCAGCAAUCAGAACCGC .(((..((((((...)))))))))((((((((....))))))..((((((((((........(((......))))))))))))).....)).. (-30.34 = -30.98 + 0.64)



| Location | 9,277,293 – 9,277,386 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -26.08 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9277293 93 - 22224390 GCGGUUCUGAUUGCUGGCGGCAUUUAACUGCAACAAUUACGCCAGCAACAUGCAACAUUAGCGCUGCCCGGCGGCAAUUACACUGCCUUACCA (((((.((((((((((((((((......)))........)))))))).........))))).)))))..(((((........)))))...... ( -30.50) >DroSec_CAF1 59723 93 - 1 GCGGUUCUGAUUGCUGGCGGCAUUUAACUGCAACAAUUACGCCAGCAACAUGCAACAUUAGCGCUGCCCGGCGGCAAUUACACUGCCUUACCA (((((.((((((((((((((((......)))........)))))))).........))))).)))))..(((((........)))))...... ( -30.50) >DroSim_CAF1 57086 93 - 1 GCGGUUCUGAUUGCUGGCGGCAUUUAACUGCAACAAUUACGCCAGCAACAUGCAACAUUAGCGCUGCCCGGCGGCAAUUACACUGCCUUACCA (((((.((((((((((((((((......)))........)))))))).........))))).)))))..(((((........)))))...... ( -30.50) >DroEre_CAF1 66209 93 - 1 GCUGUUCUGAUUGCUUGCGGCAUUUAGCUGCAACAAUUACGCCAGCAACAUGCAACAUUAGCGCUGCCCGGCGGCAAUUACACUGCCUUACCA ((.((..((((((((((((((.....)))))))......((((.(((...(((.......))).)))..))))))))))).)).))....... ( -29.30) >DroYak_CAF1 58238 93 - 1 GCGGUUCUGAUCGCUGGCGGCAUUUAACUGCAACAAUUACGCCAGCAACAUGCAACAUUAGCGCUGCCCUGUGGCAAUUACACUGCCUUGCCA (((((.(((((.((((((((((......)))........)))))))..........))))).)))))....((((((..........)))))) ( -26.00) >consensus GCGGUUCUGAUUGCUGGCGGCAUUUAACUGCAACAAUUACGCCAGCAACAUGCAACAUUAGCGCUGCCCGGCGGCAAUUACACUGCCUUACCA (((((.((((((((((((((((......)))........)))))))).........))))).)))))..(((((........)))))...... (-26.08 = -26.72 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:58 2006