| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,229,395 – 9,229,524 |
| Length | 129 |
| Max. P | 0.873618 |

| Location | 9,229,395 – 9,229,495 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.91 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -23.02 |
| Energy contribution | -23.02 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9229395 100 - 22224390 AAGUCGGGCACGUGCAGAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCU--------GGCCCCAAAAACUGAACAGUCGACCCCGUGCGC------------CAUUUUCCCC ......(((((((((.....)))))).)))(((((....((((...))))((((.(.--------((........(((....)))....)).).))))------------...))))).. ( -28.10) >DroPse_CAF1 12652 93 - 1 AACUUGGGCACGUGCAAAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCA--------GGCCCCCAGAACCC------CAUCCCCAU-CCC------------CAUCCUGCUC ..(((((((((((((.....)))))).((.(....)...((((...)))))))))))--------))............------.........-...------------.......... ( -27.20) >DroEre_CAF1 15099 98 - 1 AAGUCGGGCACGUGCAGAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCU--------GGCCCCAAAAACUGAACAGUCGACCCC-UGCGC------------CAUUUUCCC- ......(((((((((.....)))))).)))(((((....((((...))))((((...--------((........(((....))).....))-.))))------------...))))).- ( -27.42) >DroYak_CAF1 12871 100 - 1 AAGUCGGGCACGUGCAGAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCU--------GGCCCCAAAAACUGAACAGUCGACCCCAUGCGC------------CAUUUUCCCC ......(((((((((.....)))))).)))(((((....((((...))))((((..(--------((........(((....))).....))).))))------------...))))).. ( -28.52) >DroAna_CAF1 16312 100 - 1 AAGUUGGGCACGUGCAGAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCU--------GGCCCCAAAAACUGAACAGUUGGCAGAGGAGGC------------CUUCCUCCCC ..(((.(((((((((.....)))))).)))(....)....))).((((((((((...--------.))..((.....))....)))))))).(((((.------------...))))).. ( -35.20) >DroPer_CAF1 13219 113 - 1 AACUUGGGCACGUGCAAAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCAGGCACCCAGGCCCCCAGAACCC------CAUCCCCAU-CCCCAUUCCCAUUCCCAUCCUGCUC ..(((((((((((((.....)))))).((.(....)...((((...)))))))))))))....((((............------.........-..................))))... ( -27.93) >consensus AAGUCGGGCACGUGCAGAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCU________GGCCCCAAAAACUGAACAGUCGACCCCAUGCGC____________CAUCCUCCCC .....((((((((((.....)))))).((.(....)...((((...))))))))))................................................................ (-23.02 = -23.02 + 0.00)


| Location | 9,229,423 – 9,229,524 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -27.68 |
| Energy contribution | -27.84 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
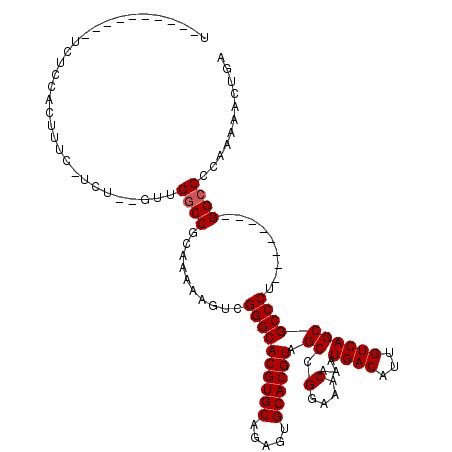
>X_DroMel_CAF1 9229423 101 - 22224390 U----------UCUCCACUUUC-UCUCAGUUGGCCGCAAAAAGUCGGGCACGUGCAGAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCU--------GGCCCCAAAAACUGA .----------...........-..((((((((((((.....)).((((((((((.....)))))).((.(....)...((((...)))))))))).--------)))).....)))))) ( -33.20) >DroPse_CAF1 12674 104 - 1 UUGUCACUGGCUGUGCGCAUU-------UUUGGCCGCAAAAACUUGGGCACGUGCAAAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCA--------GGCCCCCAGAACCC- ......((((.((((.((...-------....))))))....(((((((((((((.....)))))).((.(....)...((((...)))))))))))--------))...)))).....- ( -37.20) >DroEre_CAF1 15125 101 - 1 U----------CCUCCACUUUC-UCUCAGUUGGCCGCAAAAAGUCGGGCACGUGCAGAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCU--------GGCCCCAAAAACUGA .----------...........-..((((((((((((.....)).((((((((((.....)))))).((.(....)...((((...)))))))))).--------)))).....)))))) ( -33.20) >DroYak_CAF1 12899 101 - 1 U----------UCUCCACUUUC-UCUCAGUUGCCCGCAAAAAGUCGGGCACGUGCAGAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCU--------GGCCCCAAAAACUGA .----------...........-..((((((((((((.....).)))))((((((.....)))))).(((((.......((((...)))).....))--------)))......)))))) ( -32.30) >DroAna_CAF1 16340 99 - 1 U----------UCUGCACCUUUAGCU---UCGGCCGCAAAAAGUUGGGCACGUGCAGAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCU--------GGCCCCAAAAACUGA .----------...((.......)).---..((((((.....)).((((((((((.....)))))).((.(....)...((((...)))))))))).--------))))........... ( -29.40) >DroPer_CAF1 13253 112 - 1 UUGUCACUGGCUGUGCGCAUU-------UUUGGCCGCAAAAACUUGGGCACGUGCAAAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCAGGCACCCAGGCCCCCAGAACCC- ......((((.((....))..-------...((((.......(((((((((((((.....)))))).((.(....)...((((...)))))))))))))......)))).)))).....- ( -39.02) >consensus U__________UCUCCACUUUC_UCU__GUUGGCCGCAAAAAGUCGGGCACGUGCAGAGUGCACGUAGCCGGAAACAAAUGACAUUGUCAGCGCCCU________GGCCCCAAAAACUGA ...............................((((..........((((((((((.....)))))).((.(....)...((((...)))))))))).........))))........... (-27.68 = -27.84 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:41 2006