
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,229,033 – 9,229,174 |
| Length | 141 |
| Max. P | 0.998447 |

| Location | 9,229,033 – 9,229,140 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 92.19 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -27.51 |
| Energy contribution | -29.11 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9229033 107 + 22224390 ---AAAACGGGAGCUGGGCAAAGGAAAGGCAAAGGGAAUUCUGGCUGCCGACAACAAAAGACUGUCAUAAAAUGGCAGCUGCAAGGAUGCGCCUCAACUUUUGUGGCUGC ---......(.((((..(((((((..((((............(((((((((((.........)))).......)))))))(((....)))))))...))))))))))).) ( -32.01) >DroSec_CAF1 12309 102 + 1 --------GGGAGCUAGGCAAAGGAAAGGCAAAGGGAAUUCUGGCUGCCGACAACAAAAGACUGUCAUAAAAUGGCAGCUGCAAGGAUGCGCCUCAACUUUUGUGGCUGC --------.(.((((..(((((((..((((............(((((((((((.........)))).......)))))))(((....)))))))...))))))))))).) ( -32.01) >DroSim_CAF1 23177 102 + 1 --------GGGAGCUGGGCAAAGGAAAGGCAAAGGGAAUUCUGGCUGCCGACAACAAAAGACUGUCAUAAAAUGGCAGCUGCAAGGAUGCGCCUCAACUUUUGUGGCUGC --------.(.((((..(((((((..((((............(((((((((((.........)))).......)))))))(((....)))))))...))))))))))).) ( -32.01) >DroEre_CAF1 14739 105 + 1 AGAAAGAAGGGAGCUGGCC-A----AAGGCAAAGGGAAUUCUGGCUGCCGACAACAAAAGACUGUCAUAAAAUGGCAGCUGCAAGGAUGCGCCUCAACUUUUGUGGCUGC ........((.((((.(((-.----..)))....((....)))))).))((((.........))))........(((((..((((..(........)..))))..))))) ( -28.00) >DroYak_CAF1 12487 104 + 1 --AAAGAAGGGAGCUGGGCAA----AAGGCAAAGGGAAUUCUGGCUGCCGACAACAAAAGACUGUCAUAAAAUGGCAGCUGCAAGGAUGCGCCUCAACUUUUGUGGCUGC --.......(.((((..((((----(((....((((.((((((((((((((((.........)))).......)))))))...))))).).)))...))))))))))).) ( -30.11) >consensus ___AA_A_GGGAGCUGGGCAAAGGAAAGGCAAAGGGAAUUCUGGCUGCCGACAACAAAAGACUGUCAUAAAAUGGCAGCUGCAAGGAUGCGCCUCAACUUUUGUGGCUGC .........(.((((..(((((((..((((............(((((((((((.........)))).......)))))))(((....)))))))...))))))))))).) (-27.51 = -29.11 + 1.60)



| Location | 9,229,033 – 9,229,140 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 92.19 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -26.72 |
| Energy contribution | -27.72 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9229033 107 - 22224390 GCAGCCACAAAAGUUGAGGCGCAUCCUUGCAGCUGCCAUUUUAUGACAGUCUUUUGUUGUCGGCAGCCAGAAUUCCCUUUGCCUUUCCUUUGCCCAGCUCCCGUUUU--- ..(((....((((..((((((((....))).((((((.....(..((((....))))..).)))))).............)))))..)))).....)))........--- ( -30.50) >DroSec_CAF1 12309 102 - 1 GCAGCCACAAAAGUUGAGGCGCAUCCUUGCAGCUGCCAUUUUAUGACAGUCUUUUGUUGUCGGCAGCCAGAAUUCCCUUUGCCUUUCCUUUGCCUAGCUCCC-------- ..(((....((((..((((((((....))).((((((.....(..((((....))))..).)))))).............)))))..)))).....)))...-------- ( -30.50) >DroSim_CAF1 23177 102 - 1 GCAGCCACAAAAGUUGAGGCGCAUCCUUGCAGCUGCCAUUUUAUGACAGUCUUUUGUUGUCGGCAGCCAGAAUUCCCUUUGCCUUUCCUUUGCCCAGCUCCC-------- ..(((....((((..((((((((....))).((((((.....(..((((....))))..).)))))).............)))))..)))).....)))...-------- ( -30.50) >DroEre_CAF1 14739 105 - 1 GCAGCCACAAAAGUUGAGGCGCAUCCUUGCAGCUGCCAUUUUAUGACAGUCUUUUGUUGUCGGCAGCCAGAAUUCCCUUUGCCUU----U-GGCCAGCUCCCUUCUUUCU ((.((((........((((((((....))).((((((.....(..((((....))))..).)))))).............)))))----)-)))..))............ ( -29.30) >DroYak_CAF1 12487 104 - 1 GCAGCCACAAAAGUUGAGGCGCAUCCUUGCAGCUGCCAUUUUAUGACAGUCUUUUGUUGUCGGCAGCCAGAAUUCCCUUUGCCUU----UUGCCCAGCUCCCUUCUUU-- ...........(((((.((((((....))).((((((.....(..((((....))))..).))))))..................----..)))))))).........-- ( -27.00) >consensus GCAGCCACAAAAGUUGAGGCGCAUCCUUGCAGCUGCCAUUUUAUGACAGUCUUUUGUUGUCGGCAGCCAGAAUUCCCUUUGCCUUUCCUUUGCCCAGCUCCC_U_UU___ ..(((....((((..((((((((....))).((((((.....(..((((....))))..).)))))).............)))))..)))).....)))........... (-26.72 = -27.72 + 1.00)


| Location | 9,229,070 – 9,229,174 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 87.91 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -29.39 |
| Energy contribution | -29.41 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
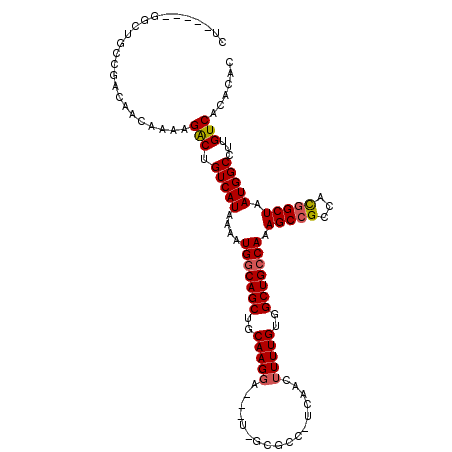
>X_DroMel_CAF1 9229070 104 + 22224390 CU-----GGCUGCCGACAACAAAAGACUGUCAUAAAAUGGCAGCUGCAAGGA---U-GCGCC-UCAACUUUUGUGGCUGCCAAAGCCGCCGCGGCUAAUGGCCUUGUCACACAC .(-----(((.(((((((.........))))......(((((((..((((..---(-.....-...)..))))..))))))).(((((...)))))...)))...))))..... ( -36.10) >DroPse_CAF1 12302 113 + 1 CUGCCGGUGGGGCAGACAACAAAAGGCUGUCAUAAAAUGGCAGCUGCAAGGCGCUCGGGCCC-UCAACUUUUGUGGCUGCCAAAGCCGCCAUGGCUAAUGGCCUUGUCACACAC .....(((..(((((...(((((((((((((((...))))))))....(((.((....))))-)...)))))))..)))))...)))(((((.....)))))............ ( -44.40) >DroEre_CAF1 14774 104 + 1 CU-----GGCUGCCGACAACAAAAGACUGUCAUAAAAUGGCAGCUGCAAGGA---U-GCGCC-UCAACUUUUGUGGCUGCCAAAGCCGCCGCGGCUAAUGGCCUUGUCAUACAC .(-----(((.(((((((.........))))......(((((((..((((..---(-.....-...)..))))..))))))).(((((...)))))...)))...))))..... ( -36.20) >DroYak_CAF1 12521 104 + 1 CU-----GGCUGCCGACAACAAAAGACUGUCAUAAAAUGGCAGCUGCAAGGA---U-GCGCC-UCAACUUUUGUGGCUGCCAAAGCCGCCGCGGCUAAUGGCCUUGUCAUACAC .(-----(((.(((((((.........))))......(((((((..((((..---(-.....-...)..))))..))))))).(((((...)))))...)))...))))..... ( -36.20) >DroAna_CAF1 15997 105 + 1 CU-----AGGAGCCGACAACAAAAGACUGUCAUAAAAUGGCAGCUGCAAGUA---U-GCGCCUUCAACUUUUGUGGCUGCCAAAGCCGCCAUUGCUAAUGGCCUUGUCACACAC ..-----.......(((((((((((.(((((((...)))))))..(((....---)-))........)))))))((((.....))))(((((.....)))))..))))...... ( -30.80) >DroPer_CAF1 12884 113 + 1 CUGCUCGUGGGGCAGACAACAAAAGGCUGUCAUAAAAUGUCAGCUGCAAGGCGCUCGGGCCC-UCAACUUUUGUGGCUGCCAAAGCCGCCAUGGCUAAUGGCCUUGUCACACAC (((((.....))))).........(((((.(((...))).)))))(((((((.(...((((.-.........(((((.((....)).)))))))))...))))))))....... ( -39.50) >consensus CU_____GGCUGCCGACAACAAAAGACUGUCAUAAAAUGGCAGCUGCAAGGA___U_GCGCC_UCAACUUUUGUGGCUGCCAAAGCCGCCACGGCUAAUGGCCUUGUCACACAC ........................(((.(((((....(((((((..(((((.................)))))..))))))).(((((...))))).)))))...)))...... (-29.39 = -29.41 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:39 2006