| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,213,052 – 9,213,212 |
| Length | 160 |
| Max. P | 0.715983 |

| Location | 9,213,052 – 9,213,172 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -21.38 |
| Energy contribution | -21.97 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9213052 120 - 22224390 AUUCAGAAGAUUUCGUAUAAUGGCGCCAAUCGCAAGAUAAUCCGCAGAGAUCUGCCCAACCCCAUGGGCAUUGCUGUCUAUCUGAACGAUGUCUACUGGGUGGAUAGGAAUCUGAUGACC ..(((..(((((((..............(((....))).((((((((....))))...((((..(((((((((...((.....)).)))))))))..)))))))).)))))))..))).. ( -41.00) >DroVir_CAF1 11144 120 - 1 AUACAGAAGAUCUCCUACAACGGCGCCAAUCGGAAAAUUAUUCGUCGCGACUUGCCCAAUCCCAUGGGCAUCGCCAUUUUCCUGAAUGAUGUUUACUGGGUAGAUCGCAAUCUUAUGACU ........(((((((((....((((.((.(((((((((........((((..((((((......)))))))))).))))))).)).)).))))...)))).))))).............. ( -32.40) >DroGri_CAF1 12388 120 - 1 AUACAGAAGAUCUCGUAUAAUGGCGUCAAUCGCAAGGUGAUCCGACGUGAUCUGCCCAAUCCGAUGGGCAUUGCCAUUUUCCGCAACGAUGUCUACUGGGUGGAUCGCAAUCUGAUGACA ...((((.............(((.((((.((....)))))))))..((((((..((((....(.(((((((((.............))))))))))))))..))))))..))))...... ( -38.32) >DroWil_CAF1 11888 120 - 1 AUACAAAAGAUAUCCUACAAUGGCGUCAAUCGCAAGGUCAUUCGUCGUGACUUACCCAAUCCCAUGGGCAUAGCAAUCUAUCUGAACGAUGUCUAUUGGGUCGAUAGGAAUUUGAUGACC ...((..((((.((((((.((((((...(((....)))....)))))).....(((((.....((((((((..((.......))....))))))))))))).).)))))))))..))... ( -28.80) >DroMoj_CAF1 12299 120 - 1 AUUCAGAAGAUAUCGUACAAUGGCGUCAAUGCCAAGGUGAUACGCCGAGACCUACCCAAUCCGGUGGGCAUUGCCAUUUAUCUGAACGAUGUGUACUGGGUGGAUGGCAAUCUCAAUUCG .....(((.....((((((.(((((....)))))...)).))))..((((((((((......))))))..((((((((((((((.((.....)).).)))))))))))))))))..))). ( -38.20) >DroAna_CAF1 12694 120 - 1 AUCCAGAAGAUCUCGUACAACGGCGCCAACCGCAAGGUCAUCCGGAGGGACCUGCCCAAUCCCAUGGGCAUUGCCGUCUACCUGAACGACGUCUACUGGGUGGACCGCAACCUGAUGACC ((((((.((((.((((.((.(((.....(((....)))...)))((.((...((((((......))))))...)).))....)).)))).)))).))))))................... ( -41.50) >consensus AUACAGAAGAUCUCGUACAAUGGCGCCAAUCGCAAGGUCAUCCGCCGAGACCUGCCCAAUCCCAUGGGCAUUGCCAUCUACCUGAACGAUGUCUACUGGGUGGAUCGCAAUCUGAUGACC ...(((.((((.((((.(((((((....(((....)))..............((((((......))))))..))))).....)).)))).)))).)))...................... (-21.38 = -21.97 + 0.59)

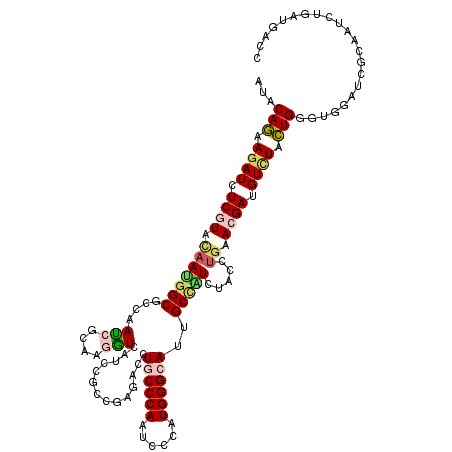

| Location | 9,213,092 – 9,213,212 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -23.17 |
| Energy contribution | -23.57 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
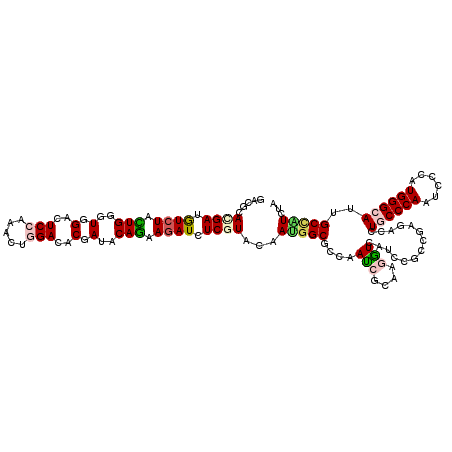
>X_DroMel_CAF1 9213092 120 - 22224390 GACCCAUGACAUCUACUGGGUGGACUCGAAACUGGACACCAUUCAGAAGAUUUCGUAUAAUGGCGCCAAUCGCAAGAUAAUCCGCAGAGAUCUGCCCAACCCCAUGGGCAUUGCUGUCUA (((..((((.((((.((((((((..((.......))..)))))))).)))).))))......(((...(((....)))....)))..((...((((((......))))))...))))).. ( -36.20) >DroVir_CAF1 11184 120 - 1 GACGCAUGAUGUCUUUUGGGUGGACUCGAAACUGGAUACGAUACAGAAGAUCUCCUACAACGGCGCCAAUCGGAAAAUUAUUCGUCGCGACUUGCCCAAUCCCAUGGGCAUCGCCAUUUU ((((.(((((...(((((..(((..(((.....(((...(((.......)))))).....)))..)))..))))).))))).))))((((..((((((......))))))))))...... ( -34.20) >DroGri_CAF1 12428 120 - 1 GACGCACGAUGUGUUUUGGGUUGACUCCAAGCUGGACACCAUACAGAAGAUCUCGUAUAAUGGCGUCAAUCGCAAGGUGAUCCGACGUGAUCUGCCCAAUCCGAUGGGCAUUGCCAUUUU .....((((.((.(((((...((..(((.....)))...))..))))).)).))))..(((((((((.(((((...)))))..)))......((((((......))))))..)))))).. ( -35.10) >DroWil_CAF1 11928 120 - 1 AACCCAUGAUGUCUAUUGGGUUGAUUCCAAAUUGGAUACGAUACAAAAGAUAUCCUACAAUGGCGUCAAUCGCAAGGUCAUUCGUCGUGACUUACCCAAUCCCAUGGGCAUAGCAAUCUA ..((((((......(((((((((((.(((....((((((.........).))))).....))).))))......(((((((.....))))))))))))))..))))))............ ( -31.80) >DroMoj_CAF1 12339 120 - 1 AACGCACGAUGUCUACUGGGUGGACUCCAAGCUGGAUACGAUUCAGAAGAUAUCGUACAAUGGCGUCAAUGCCAAGGUGAUACGCCGAGACCUACCCAAUCCGGUGGGCAUUGCCAUUUA ...(.(((((((((.((((((.(..(((.....)))..).)))))).))))))))).)(((((((...(((((..((((...))))......((((......)))))))))))))))).. ( -43.60) >DroAna_CAF1 12734 120 - 1 GACGCACGACAUCUUCUGGGUGGACUCCAAGCUGGACACGAUCCAGAAGAUCUCGUACAACGGCGCCAACCGCAAGGUCAUCCGGAGGGACCUGCCCAAUCCCAUGGGCAUUGCCGUCUA ((((.((((.(((((((((((.(..(((.....)))..).))))))))))).)))).)...(((....(((....)))..(((....)))..((((((......))))))..)))))).. ( -50.90) >consensus GACGCACGAUGUCUACUGGGUGGACUCCAAACUGGACACGAUACAGAAGAUCUCGUACAAUGGCGCCAAUCGCAAGGUCAUCCGCCGAGACCUGCCCAAUCCCAUGGGCAUUGCCAUCUA .....((((.((((.(((..(.(..(((.....)))..).)..))).)))).))))...(((((....(((....)))..............((((((......))))))..)))))... (-23.17 = -23.57 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:35 2006