
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,185,679 – 9,185,893 |
| Length | 214 |
| Max. P | 0.949041 |

| Location | 9,185,679 – 9,185,792 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.13 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -19.05 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9185679 113 + 22224390 UUCCGCCUUUCCC-UGAUGCCCUUAACUUCAUCAUCAUCGGCAAUCCUUCGCUGCAUUAUAAUUGGCAGAAUGACCUCAGACUGUCAGUUAUCC------AUUUCCAUUUCCAUUUUCUU ....(((......-(((((..........))))).....)))................(((((((((((..((....))..)))))))))))..------.................... ( -19.70) >DroSec_CAF1 13357 113 + 1 UUCCGCCUUUCCC-UGAUGACUUUAACUUCAUCAUCGUCGGCAGUCCUUCGCUGCAUUAUAAUUGGCAGAAUGACCUCAGACUGUCAGUUAUCC------AUUUCCAUUUCCAUUUUCUU .............-((((((........))))))......(((((.....)))))...(((((((((((..((....))..)))))))))))..------.................... ( -24.30) >DroEre_CAF1 11158 120 + 1 UGCUGCCUUUCCCUUGAUGCCUUCGACUUCAUCAUCGUCGGCAGUCCUUCGUUGCAUUAUAAUUGGCAGAAUGACCUCAGACUGUCAGUUAUCCAUGUCCAUUUCCAUUUCCAUUUUCUU .((((((........((.....))(((.........)))))))))........((((.(((((((((((..((....))..)))))))))))..))))...................... ( -26.40) >DroYak_CAF1 11342 113 + 1 UUCUGCCUUUUCC-GGAUGCCUUCAACUUCAUCAUCGUCGGCAGUCCUUCGCUGCACUAUAAUUGGCAGAAUGACCUCAGACUGUCAGUUAUCC------GUUUCCAUUUCCAUUUUCUU .............-(((.((....................(((((.....)))))...(((((((((((..((....))..)))))))))))..------)).))).............. ( -24.40) >consensus UUCCGCCUUUCCC_UGAUGCCUUCAACUUCAUCAUCGUCGGCAGUCCUUCGCUGCAUUAUAAUUGGCAGAAUGACCUCAGACUGUCAGUUAUCC______AUUUCCAUUUCCAUUUUCUU ....(((.......(((((..........))))).....)))................(((((((((((..((....))..)))))))))))............................ (-19.05 = -19.30 + 0.25)



| Location | 9,185,718 – 9,185,829 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 93.42 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -20.70 |
| Energy contribution | -21.33 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9185718 111 + 22224390 GCAAUCCUUCGCUGCAUUAUAAUUGGCAGAAUGACCUCAGACUGUCAGUUAUCC------AUUUCCAUUUCCAUUUUCUUUGCUUUUUAUUUAGCUGCCAGUUAGUUUCGCACUGGC (((.........)))...(((((((((((..((....))..)))))))))))..------.....................(((........))).((((((.........)))))) ( -23.10) >DroSec_CAF1 13396 111 + 1 GCAGUCCUUCGCUGCAUUAUAAUUGGCAGAAUGACCUCAGACUGUCAGUUAUCC------AUUUCCAUUUCCAUUUUCUUUGCCUUUUAUUUAGCUGCCAGUUAGUUUCGCACUGGC (((((.....)))))...(((((((((((..((....))..)))))))))))..------....................................((((((.........)))))) ( -26.20) >DroEre_CAF1 11198 117 + 1 GCAGUCCUUCGUUGCAUUAUAAUUGGCAGAAUGACCUCAGACUGUCAGUUAUCCAUGUCCAUUUCCAUUUCCAUUUUCUUUGCUUUUUAUUUAGCUGCCAGUUAGUUUCGCACUCGC ((.(..((.....(((..(((((((((((..((....))..))))))))))).............................(((........)))))).....))..).))...... ( -19.90) >DroYak_CAF1 11381 111 + 1 GCAGUCCUUCGCUGCACUAUAAUUGGCAGAAUGACCUCAGACUGUCAGUUAUCC------GUUUCCAUUUCCAUUUUCUUUGCUUUUUAUUUUGCUGCCAGUUAGUUUAGCACCGGC (((((.....)))))...(((((((((((..((....))..)))))))))))((------(....................((..........))(((.((.....)).))).))). ( -22.60) >consensus GCAGUCCUUCGCUGCAUUAUAAUUGGCAGAAUGACCUCAGACUGUCAGUUAUCC______AUUUCCAUUUCCAUUUUCUUUGCUUUUUAUUUAGCUGCCAGUUAGUUUCGCACUGGC (((((.....)))))...(((((((((((..((....))..)))))))))))............................................((((((.........)))))) (-20.70 = -21.33 + 0.62)

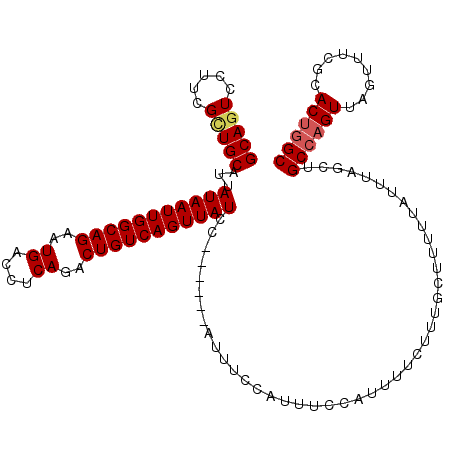
| Location | 9,185,758 – 9,185,868 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -17.68 |
| Energy contribution | -18.75 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9185758 110 + 22224390 ACUGUCAGUUAUCC------AUUUCCAUUUCCAUUUUCUUUGCUUUUUAUUUAGCUGCCAGUUAGUUUCGCACUGGCUCUCUGGACAGUUUUCCCC-GGAGCCGAAUGGAUUUCCUU .......(..((((------((((.................(((........))).((((((.........)))))).((((((..........))-))))..))))))))..)... ( -24.50) >DroSec_CAF1 13436 110 + 1 ACUGUCAGUUAUCC------AUUUCCAUUUCCAUUUUCUUUGCCUUUUAUUUAGCUGCCAGUUAGUUUCGCACUGGCUCUCUGGACAGUUUUCCCC-GGAGUCGAAUGGAUUUCCUC .......(..((((------((((................................((((((.........)))))).((((((..........))-))))..))))))))..)... ( -23.20) >DroEre_CAF1 11238 117 + 1 ACUGUCAGUUAUCCAUGUCCAUUUCCAUUUCCAUUUUCUUUGCUUUUUAUUUAGCUGCCAGUUAGUUUCGCACUCGCUCUCUGGACAAUUUUCCCCUGAAGUCGCAUGGAUUUCCAU .......(..((((((((..(((((................(((........)))((((((..(((.........)))..)))).))..........))))).))))))))..)... ( -21.80) >DroYak_CAF1 11421 111 + 1 ACUGUCAGUUAUCC------GUUUCCAUUUCCAUUUUCUUUGCUUUUUAUUUUGCUGCCAGUUAGUUUAGCACCGGCUCUCUGGACACUUUUCCCCCGAAGUCGAAUGGAUUUUCUU ......((..((((------(((.(.(((((..........((..........))((((((..((((.......))))..)))).))..........))))).))))))))...)). ( -18.10) >consensus ACUGUCAGUUAUCC______AUUUCCAUUUCCAUUUUCUUUGCUUUUUAUUUAGCUGCCAGUUAGUUUCGCACUGGCUCUCUGGACAGUUUUCCCC_GAAGUCGAAUGGAUUUCCUU .............................(((((((.....(((((......(((((((((..((((.......))))..)))).))))).......))))).)))))))....... (-17.68 = -18.75 + 1.06)


| Location | 9,185,758 – 9,185,868 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -25.44 |
| Consensus MFE | -18.74 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9185758 110 - 22224390 AAGGAAAUCCAUUCGGCUCC-GGGGAAAACUGUCCAGAGAGCCAGUGCGAAACUAACUGGCAGCUAAAUAAAAAGCAAAGAAAAUGGAAAUGGAAAU------GGAUAACUGACAGU .((...(((((((....(((-(((.....)).((((....((((((.........)))))).(((........)))........))))..)))))))------))))..))...... ( -26.20) >DroSec_CAF1 13436 110 - 1 GAGGAAAUCCAUUCGACUCC-GGGGAAAACUGUCCAGAGAGCCAGUGCGAAACUAACUGGCAGCUAAAUAAAAGGCAAAGAAAAUGGAAAUGGAAAU------GGAUAACUGACAGU .((...(((((((....(((-(((.....)).((((....((((((.........)))))).(((........)))........))))..)))))))------))))..))...... ( -26.30) >DroEre_CAF1 11238 117 - 1 AUGGAAAUCCAUGCGACUUCAGGGGAAAAUUGUCCAGAGAGCGAGUGCGAAACUAACUGGCAGCUAAAUAAAAAGCAAAGAAAAUGGAAAUGGAAAUGGACAUGGAUAACUGACAGU ((((....))))...(((((((........((((((....((.(((.........))).)).(((........)))....................)))))).......)))).))) ( -24.76) >DroYak_CAF1 11421 111 - 1 AAGAAAAUCCAUUCGACUUCGGGGGAAAAGUGUCCAGAGAGCCGGUGCUAAACUAACUGGCAGCAAAAUAAAAAGCAAAGAAAAUGGAAAUGGAAAC------GGAUAACUGACAGU ......((((.(((..(....)..)))..((.((((....((((((.........)))))).((..........))..............)))).))------)))).......... ( -24.50) >consensus AAGGAAAUCCAUUCGACUCC_GGGGAAAACUGUCCAGAGAGCCAGUGCGAAACUAACUGGCAGCUAAAUAAAAAGCAAAGAAAAUGGAAAUGGAAAU______GGAUAACUGACAGU .......((((((...((....))........((((....((((((.........)))))).(((........)))........))))))))))....................... (-18.74 = -19.18 + 0.44)

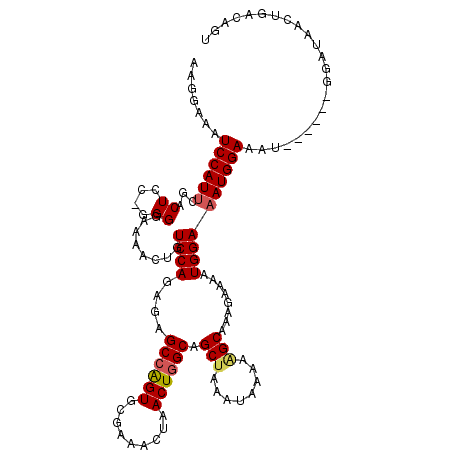
| Location | 9,185,792 – 9,185,893 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 84.80 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -15.82 |
| Energy contribution | -16.20 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9185792 101 - 22224390 UCUUUUGCUGCUCGAAAAAAA-AAAGAAGGAAAUCCAUUCGGCUCC-GGGGAAAACUGUCCAGAGAGCCAGUGCGAAACUAACUGGCAGCUAAAUAAAAAGCA ......((((((.(.......-...(((((....)).)))((((((-..(((......))).).)))))(((.....)))..).))))))............. ( -26.60) >DroSec_CAF1 13470 97 - 1 UCUUUUGCUGCUCGGAA-----AAAGGAGGAAAUCCAUUCGACUCC-GGGGAAAACUGUCCAGAGAGCCAGUGCGAAACUAACUGGCAGCUAAAUAAAAGGCA (((((......(((((.-----...(((((....)).)))...)))-))(((......))))))))((((((.........)))))).(((........))). ( -25.20) >DroEre_CAF1 11278 96 - 1 UCUGCUGCUGCUCA-------GAAAAAUGGAAAUCCAUGCGACUUCAGGGGAAAAUUGUCCAGAGAGCGAGUGCGAAACUAACUGGCAGCUAAAUAAAAAGCA ...((((((((((.-------(((..((((....)))).....)))...(((......)))...)))).(((.....)))....))))))............. ( -24.20) >DroYak_CAF1 11455 103 - 1 UCUUCUGCUGCACGAAAAAAAAAAAAAAGAAAAUCCAUUCGACUUCGGGGGAAAAGUGUCCAGAGAGCCGGUGCUAAACUAACUGGCAGCAAAAUAAAAAGCA ..(((((..((((...............(......).(((..(....)..)))..)))).))))).((((((.........)))))).((..........)). ( -20.30) >consensus UCUUCUGCUGCUCGAAA_____AAAAAAGGAAAUCCAUUCGACUCC_GGGGAAAACUGUCCAGAGAGCCAGUGCGAAACUAACUGGCAGCUAAAUAAAAAGCA .....((((.(((...............((....)).............(((......))).))).((((((.........))))))............)))) (-15.82 = -16.20 + 0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:30 2006