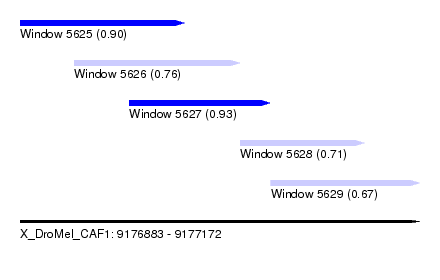
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,176,883 – 9,177,172 |
| Length | 289 |
| Max. P | 0.933350 |

| Location | 9,176,883 – 9,177,002 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 72.43 |
| Mean single sequence MFE | -24.25 |
| Consensus MFE | -13.66 |
| Energy contribution | -13.50 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9176883 119 + 22224390 GUUGUAUAUACAUAUGUAUGUAGUAUAUAGCAACGGGGUUAUUGACCCACAGGGCAUCUCUUUCUUUUUCGCCCUCUAUAUCUAUCCCUUUUCUUUCUACUUUUCACGAGUGCGAAAGG (((((((((((((....)))).)))))))))...(((((....((.....(((((...............))))).....)).)))))....((((((((((.....))))).))))). ( -30.46) >DroSec_CAF1 4769 101 + 1 ---GUA------------U---GUAUAUAGCAACGGGGUUAUUGACCCACAGGGCAUCUCUUUCUUUUUCGCCCUCUAUAUCUAUCCCUUUUCUUUCUACUUUUCAAGAGUGCGAAAGG ---..(------------(---(.((((((.....(((((...)))))..(((((...............))))))))))).))).......((((((((((.....))))).))))). ( -22.76) >DroSim_CAF1 4775 101 + 1 ---GUA------------U---GUAUAUAGCAACGGGGUUAUUGACCCACAGGGCAUCUCUUUCUUUUUCGCCCUCUAUAGCUAUCCCUUUUCUUUCUACUUUUCAAGAGUGCGAAAGG ---...------------.---....(((((....(((((...)))))..(((((...............))))).....))))).......((((((((((.....))))).))))). ( -24.76) >DroYak_CAF1 2858 103 + 1 ---GUA------------C-UCGUGUAUGGAACUGGGGUUAUUGACCCACAGGGCAUCUCUUUCUUUUUCGCCCUCUAUAUCCAUCCCUUUUCAUUCUACUUAUCAAGUGUGCGAAGGA ---...------------.-..(..((((((..(((((((...))))).))((((...............))))))))))..)...(((((.(((.((........)).))).))))). ( -25.36) >DroAna_CAF1 3314 91 + 1 ---ACA------------U-----------UCGAGGGGUUAAUGACCCUCAGGCCAUCUCUUUUACUUUCAAUUUCUACUGCCAUCUCGCUUUCUUUUGCUUA--AAGGCCACCACGGA ---...------------.-----------..(((((........))))).((((..............((........)).......((........))...--..))))........ ( -17.90) >consensus ___GUA____________U___GUAUAUAGCAACGGGGUUAUUGACCCACAGGGCAUCUCUUUCUUUUUCGCCCUCUAUAUCUAUCCCUUUUCUUUCUACUUUUCAAGAGUGCGAAAGG ...................................(((((...)))))..(((((...............)))))................(((((((((((.....))))).)))))) (-13.66 = -13.50 + -0.16)

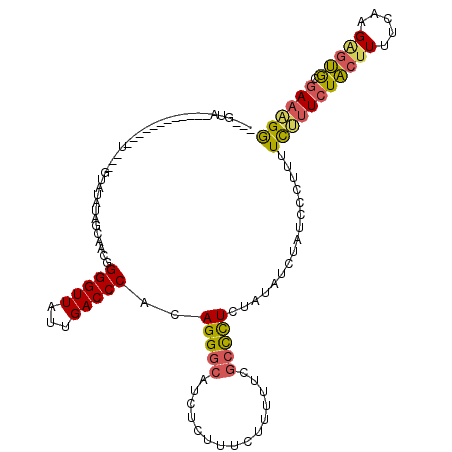
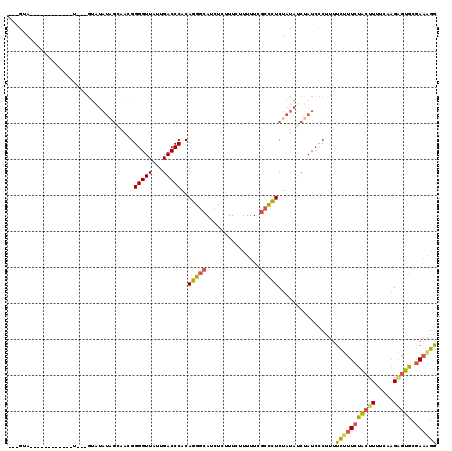
| Location | 9,176,922 – 9,177,042 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.10 |
| Mean single sequence MFE | -28.81 |
| Consensus MFE | -17.21 |
| Energy contribution | -18.05 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9176922 120 + 22224390 UAUUGACCCACAGGGCAUCUCUUUCUUUUUCGCCCUCUAUAUCUAUCCCUUUUCUUUCUACUUUUCACGAGUGCGAAAGGGAAGGCGACAGAGUGGAAGUAUCCAAAUAUACAGAUUACA .......(((((((((...............))))).....(((.((((((..((((((((((.....))))).)))))..)))).)).)))))))..((((......))))........ ( -34.76) >DroSec_CAF1 4790 119 + 1 UAUUGACCCACAGGGCAUCUCUUUCUUUUUCGCCCUCUAUAUCUAUCCCUUUUCUUUCUACUUUUCAAGAGUGCGAAAGGGAAGGCGAAAGAGUG-AAGUAUCCUAAUUUACGGAUUAUA ........((((((((...............))))).....(((.((((((..((((((((((.....))))).)))))..)))).)).))))))-....((((........)))).... ( -33.26) >DroSim_CAF1 4796 119 + 1 UAUUGACCCACAGGGCAUCUCUUUCUUUUUCGCCCUCUAUAGCUAUCCCUUUUCUUUCUACUUUUCAAGAGUGCGAAAGGGAAGGCGACAGAGUG-AAGUAUCCUAAUUUACGGAUUAUA ........((((((((...............)))))......((.((((((..((((((((((.....))))).)))))..)))).)).)).)))-....((((........)))).... ( -31.56) >DroEre_CAF1 2668 118 + 1 UAUUGACCCACAGGGCAUCUCUUUCUUUUUCGCCCUCUAUAUCCAUCUCGUUUCUUUCUACUCAU-AAGAGUGCGAAAGGGAAGGCGCCAGAGGG-AAUUAUCCUAGUUUACAGAUAUUA ......(((....(((....(((((((((((((.((((...........................-.)))).))))))))))))).)))...)))-........................ ( -28.19) >DroYak_CAF1 2881 119 + 1 UAUUGACCCACAGGGCAUCUCUUUCUUUUUCGCCCUCUAUAUCCAUCCCUUUUCAUUCUACUUAUCAAGUGUGCGAAGGAGAAGGCGACAGAGUG-AAGUAUCCUAGUUCACAGAUAUUA ((((.....(((((..(..((...((((.(((((.(((.........(((((.(((.((........)).))).)))))))).))))).)))).)-)..)..))).)).....))))... ( -26.20) >DroAna_CAF1 3327 113 + 1 UAAUGACCCUCAGGCCAUCUCUUUUACUUUCAAUUUCUACUGCCAUCUCGCUUUCUUUUGCUUA--AAGGCCACCACGGAGCGAGGGAGAGAGCA-AAAUA----AAUUCUUAAAUAAUA ...((..((((.(((..........................))).(((((((((.....(((..--..)))......)))))))))))).)..))-.....----............... ( -18.87) >consensus UAUUGACCCACAGGGCAUCUCUUUCUUUUUCGCCCUCUAUAUCCAUCCCUUUUCUUUCUACUUAUCAAGAGUGCGAAAGGGAAGGCGACAGAGUG_AAGUAUCCUAAUUUACAGAUAAUA ...((.(((...)))))((.(((((((((((((.((((.............................)))).))))))))))))).))................................ (-17.21 = -18.05 + 0.84)
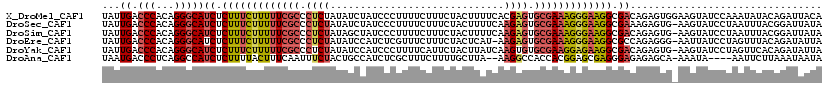
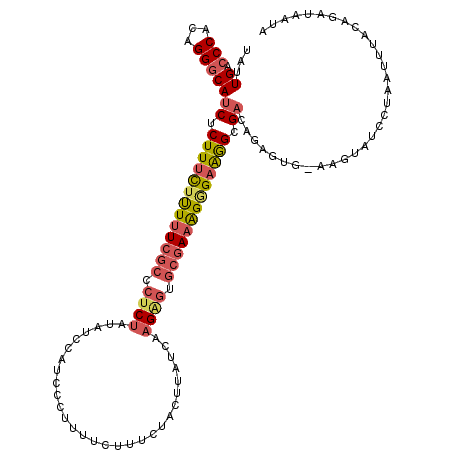
| Location | 9,176,962 – 9,177,064 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.46 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -12.38 |
| Energy contribution | -12.58 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9176962 102 + 22224390 AUCUAUCCCUUUUCUUUCUACUUUUCACGAGUGCGAAAGGGAAGGCGACAGAGUGGAAGUAUCCAAAUAUACAGAUUACAAUCCCAGCGAAU----------------ACACAUAUAG-- .(((.((((((..((((((((((.....))))).)))))..)))).)).)))(((...((((......)))).(((....))).........----------------.)))......-- ( -24.50) >DroSec_CAF1 4830 107 + 1 AUCUAUCCCUUUUCUUUCUACUUUUCAAGAGUGCGAAAGGGAAGGCGAAAGAGUG-AAGUAUCCUAAUUUACGGAUUAUAAUCCCAGCGAAU----AUA--------UACACAUAUAUUU .(((.((((((..((((((((((.....))))).)))))..)))).)).)))(((-((.........)))))((((....))))....((((----(((--------(....)))))))) ( -28.50) >DroSim_CAF1 4836 109 + 1 AGCUAUCCCUUUUCUUUCUACUUUUCAAGAGUGCGAAAGGGAAGGCGACAGAGUG-AAGUAUCCUAAUUUACGGAUUAUAAUCCCAGCGAAUU--UAUA--------UACACAUAUAUAU .(((.((((((..((((((((((.....))))).)))))..)))).))....(((-((.........)))))((((....)))).))).....--((((--------((....)))))). ( -26.60) >DroEre_CAF1 2708 110 + 1 AUCCAUCUCGUUUCUUUCUACUCAU-AAGAGUGCGAAAGGGAAGGCGCCAGAGGG-AAUUAUCCUAGUUUACAGAUAUUAUUCCCAGCAAAUAGCUACA--------UACAUAUAUGUAC .....(((.((.(((((((((((..-..)))))(....))))))).)).)))(((-(((((((..........))))..))))))(((.....)))(((--------((....))))).. ( -25.40) >DroYak_CAF1 2921 119 + 1 AUCCAUCCCUUUUCAUUCUACUUAUCAAGUGUGCGAAGGAGAAGGCGACAGAGUG-AAGUAUCCUAGUUCACAGAUAUUAAUUCCAGCGAAUAUCUAUAUAACUACACAUACAUACAUAC .((..(((((((((.((((((.........))).))).))))))).))..))(((-((.((...)).)))))(((((((..........)))))))........................ ( -21.60) >DroAna_CAF1 3367 104 + 1 UGCCAUCUCGCUUUCUUUUGCUUA--AAGGCCACCACGGAGCGAGGGAGAGAGCA-AAAUA----AAUUCUUAAAUAAUAUUU------AAUCCCUUU---ACUAGAAGCUCUAAUUUAA .....(((((((((.....(((..--..)))......)))))))))...(((((.-.....----.....((((((...))))------))...((..---...))..)))))....... ( -18.70) >consensus AUCCAUCCCUUUUCUUUCUACUUAUCAAGAGUGCGAAAGGGAAGGCGACAGAGUG_AAGUAUCCUAAUUUACAGAUAAUAAUCCCAGCGAAU___UAUA________UACACAUAUAUAC .....((((((((((((((((((.....))))).))))))))))).))........................................................................ (-12.38 = -12.58 + 0.20)


| Location | 9,177,042 – 9,177,132 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 61.81 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -10.26 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9177042 90 + 22224390 AUCCCAGCGAAU----------------ACACAUAUAG-----------UUUUAGUUUUAAUAGAAGUCCUUGGAAUUUACAAGCCUACAAGGAAACUACAAUAGGCGAUAAGCCCA ......((....----------------.........(-----------(...((((((((.........)))))))).))..(((((..((....))....))))).....))... ( -13.10) >DroEre_CAF1 2786 108 + 1 UUCCCAGCAAAUAGCUACA--------UACAUAUAUGUACAUACUUAGUUUUUAAUUUUGUAAUGAGUUGUUUGGACUGACACGCCUACAAGAAACCUGCA-UAGGCGGAAAGCUUA (((((((((((((((((((--------((....))))).(((((..(((.....)))..)).))))))))))))..)))....(((((..((....))...-)))))))))...... ( -23.70) >DroYak_CAF1 3000 117 + 1 AUUCCAGCGAAUAUCUAUAUAACUACACAUACAUACAUACAUAUAUAGCUUUCAAUUUCGUAAUGAGUUCUUUGGACUCACACGCCUACAAGGAAACUAUAGUGGGCGGCAAACUUA ......(((((...(((((((....................)))))))..)))..........((((((.....))))))..(((((((.((....))...)))))))))....... ( -27.45) >consensus AUCCCAGCGAAUA_CUA_A_________ACACAUAUAUACAUA__UAG_UUUUAAUUUUGUAAUGAGUUCUUUGGACUCACACGCCUACAAGGAAACUACA_UAGGCGGAAAGCUUA ................................................................(((((.....)))))...((((((..((....))....))))))......... (-10.26 = -10.60 + 0.34)

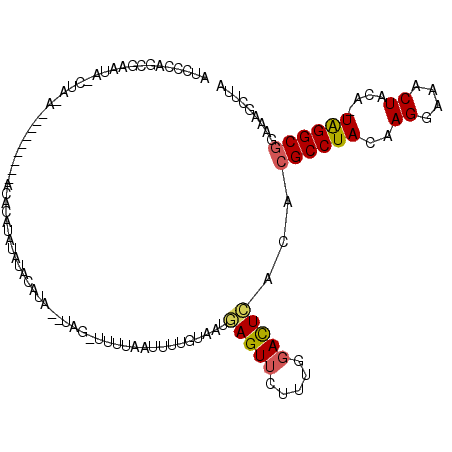

| Location | 9,177,064 – 9,177,172 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.86 |
| Mean single sequence MFE | -21.31 |
| Consensus MFE | -1.70 |
| Energy contribution | -2.18 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.08 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9177064 108 + 22224390 ---------UUUUAGUUUUAAUAGAAGUCCUUGGAAUUUACAAGCCUACAAGGAAACUACAAUAGGCGAUAAGCCCAUAAAGCAAAUAAACAAACUAACGGUAAAGGAAACGCUUGA ---------.....((((((.(((...((((((...............))))))..))).....(((.....)))..))))))......................(....)...... ( -18.16) >DroSec_CAF1 4937 95 + 1 AUAG----UUUUUAGUUUUAAUAGGAAUCCUUGG------------------GAAACUACAAUGGGCGGUAAUCCCAUAAAGCAAAUAAACAAACUAACGGUAAAGCAAACGCUUGA .(((----(((.(((((((...(((...)))...------------------)))))))..(((((.......))))).............))))))......((((....)))).. ( -18.00) >DroSim_CAF1 4945 99 + 1 AUAUUUGUUUUUUAGUAUUAAUAGGAAUCCUUGG------------------GAAACUACAAUAGGCGGUAAUCCCAUAAAGCAAAUAAACAAACUAACGGUAAAGCAAACGCUUGA .((((((((((...........(((...)))(((------------------((.((..(.....)..))..))))).))))))))))...............((((....)))).. ( -16.40) >DroEre_CAF1 2818 116 + 1 AUACUUAGUUUUUAAUUUUGUAAUGAGUUGUUUGGACUGACACGCCUACAAGAAACCUGCA-UAGGCGGAAAGCUUAUAAAGCAAAUAAACAAACUAACGGUAAAGCUAAACCUUGA ....(((((((.....(((((.((((((((((......))).((((((..((....))...-))))))...)))))))...))))).....))))))).(((........))).... ( -24.20) >DroYak_CAF1 3040 117 + 1 AUAUAUAGCUUUCAAUUUCGUAAUGAGUUCUUUGGACUCACACGCCUACAAGGAAACUAUAGUGGGCGGCAAACUUAUAAAGCAAAUAAACAAACUAACGGUAAAGCUAAAGCUUGA .....(((((((.....((((..((((((.....))))))..(((((((.((....))...))))))).............................)))).)))))))........ ( -29.80) >consensus AUAU_U__UUUUUAGUUUUAAUAGGAGUCCUUGG_A_U_ACA_GCCUACAAGGAAACUACAAUAGGCGGUAAGCCCAUAAAGCAAAUAAACAAACUAACGGUAAAGCAAACGCUUGA .............................................................(((((.......))))).........................((((....)))).. ( -1.70 = -2.18 + 0.48)


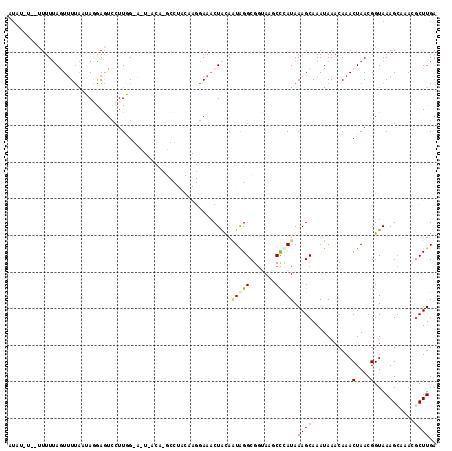
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:26 2006