
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,167,307 – 9,167,519 |
| Length | 212 |
| Max. P | 0.800808 |

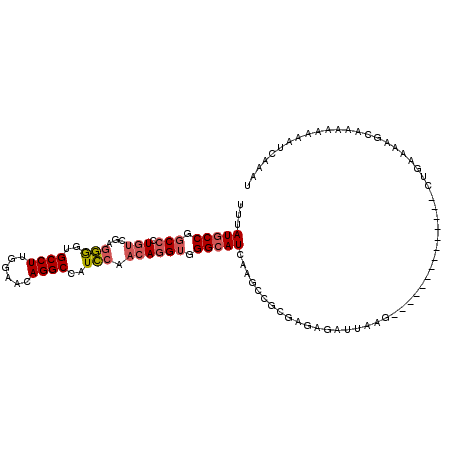
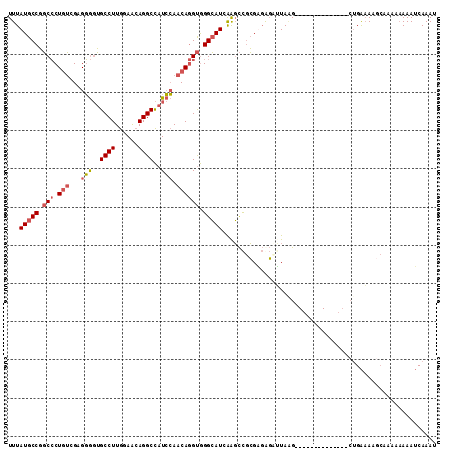
| Location | 9,167,307 – 9,167,405 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 68.41 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -15.99 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
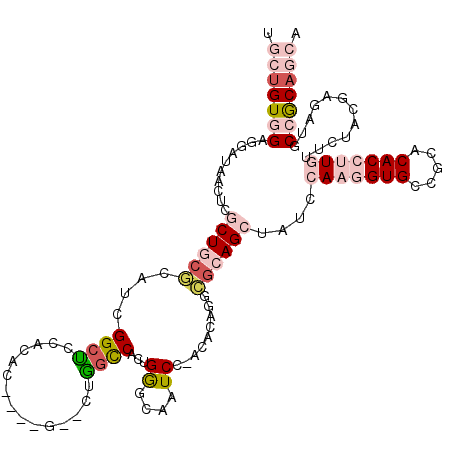
>X_DroMel_CAF1 9167307 98 + 22224390 UUUAUGCCGGCCCUGUCGAGGGGUGCCUUGGAACAGGCCAUCCAACAGGUGGGCAUAAAGCCGCGAGAGAUUAAG--------------CUGAAAAGCAAAAAAAAUCAAAU ((((((((.(((.(((....((((((((......))).))))).)))))).))))))))..(....).......(--------------((....))).............. ( -33.00) >DroVir_CAF1 86487 107 + 1 UUUAUGCCUUCACUUACGCGUACGGCCUUAGAGCAGGCUAUAAAGCAAGCAGGAAUUGAUUUUAGUUGUAAUAUGGGUCAAACCUGCGACUGGAA-----CCAAAAACAAAU ......(((...(((.(...((..((((......))))..))..).))).)))..(((.(((((((((((....((......)))))))))))))-----.)))........ ( -23.00) >DroSim_CAF1 1994 98 + 1 UUUAUGCCGGCCCUGUCGAGGGGUGCCUUGGAACAGGCCAUCCAACAGGUGGGCAUCAGGCCGCGAGAGAUUAGG--------------CUGAAAAGCAAAAAAAAUCAAAU ....((((.(((.(((....((((((((......))).))))).)))))).))))((((.((.(....)....))--------------))))................... ( -31.30) >DroYak_CAF1 1987 98 + 1 UUUAUGCCGGCCCUGUCGCGGGGUGCCUUGGAACAGGCCAUCCAACAGGUGGGCAUCAAGCCGCGUGGGCGUAAG--------------GCGACAAACAAAGCAAAUCAAAU ....((((.((((...((((((((((((((((........))))......)))))))...))))).))))....)--------------))).................... ( -34.40) >consensus UUUAUGCCGGCCCUGUCGAGGGGUGCCUUGGAACAGGCCAUCCAACAGGUGGGCAUCAAGCCGCGAGAGAUUAAG______________CUGAAAAGCAAAAAAAAUCAAAU ...(((((.(((.(((...(((..((((......))))..))).)))))).)))))........................................................ (-15.99 = -16.93 + 0.94)

| Location | 9,167,307 – 9,167,405 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 68.41 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -18.62 |
| Energy contribution | -17.25 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9167307 98 - 22224390 AUUUGAUUUUUUUUGCUUUUCAG--------------CUUAAUCUCUCGCGGCUUUAUGCCCACCUGUUGGAUGGCCUGUUCCAAGGCACCCCUCGACAGGGCCGGCAUAAA ....((((......(((....))--------------)..))))..........(((((((..((((((((...((((......)))).....))))))))...))))))). ( -29.20) >DroVir_CAF1 86487 107 - 1 AUUUGUUUUUGG-----UUCCAGUCGCAGGUUUGACCCAUAUUACAACUAAAAUCAAUUCCUGCUUGCUUUAUAGCCUGCUCUAAGGCCGUACGCGUAAGUGAAGGCAUAAA ........(((.-----...)))..(((((.((((..................))))..))))).((((((...((((......))))....(((....))))))))).... ( -24.37) >DroSim_CAF1 1994 98 - 1 AUUUGAUUUUUUUUGCUUUUCAG--------------CCUAAUCUCUCGCGGCCUGAUGCCCACCUGUUGGAUGGCCUGUUCCAAGGCACCCCUCGACAGGGCCGGCAUAAA ...................((((--------------((...........)).))))((((..((((((((...((((......)))).....))))))))...)))).... ( -28.40) >DroYak_CAF1 1987 98 - 1 AUUUGAUUUGCUUUGUUUGUCGC--------------CUUACGCCCACGCGGCUUGAUGCCCACCUGUUGGAUGGCCUGUUCCAAGGCACCCCGCGACAGGGCCGGCAUAAA ....(((..((...))..)))((--------------(....((((.((((((.....)))........((...((((......))))...)))))...)))).)))..... ( -30.20) >consensus AUUUGAUUUUUUUUGCUUUUCAG______________CUUAAUCCCACGCGGCUUGAUGCCCACCUGUUGGAUGGCCUGUUCCAAGGCACCCCGCGACAGGGCCGGCAUAAA ..................................................(((.....)))....((((((...((((......))))..(((......))))))))).... (-18.62 = -17.25 + -1.38)

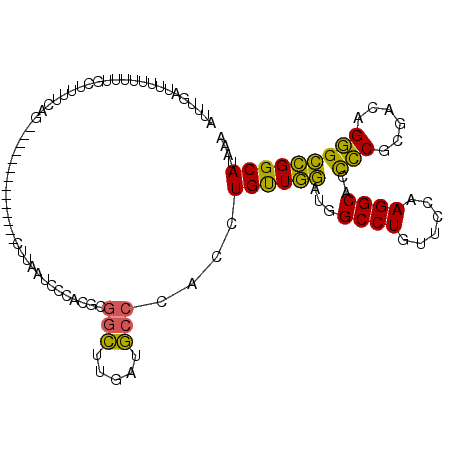
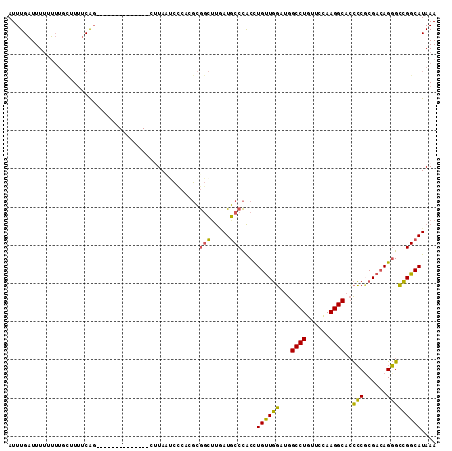
| Location | 9,167,405 – 9,167,519 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.66 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -16.40 |
| Energy contribution | -19.40 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9167405 114 + 22224390 UGCUGUGGAGGAUAACUCGCUGCGGAUCGGCUCCACCC----G--UUGGCCACUGGGCAAUCCGACUCAGGCGCAGCUAUCCAAGGUGCCGCACACCUUGUUCUACGAGAUGCCGCAGCA .(((((((.(((((....((((((....((((......----.--..)))).(((((........))))).)))))))))))((((((.....)))))).............))))))). ( -45.50) >DroVir_CAF1 86594 114 + 1 CAGCGUAGUGCACAACAAACUACUCAUUGGCAAAACUCUGAUGCAGUUGCCA--GAGAAAUC----ACUGGAGCAGCAAUCGAAGGUGCCUAGCACCCUGUUCUACGAAAUGCCACAACA ..(.(((((.........))))).)..(((((...(((((..((....))))--))).....----..((((((((........(((((...))))))))))))).....)))))..... ( -32.40) >DroSim_CAF1 2092 114 + 1 UGCUGUGGAGGAUAACUCGCUGCGGAUCGGCUCCACCC----G--CUGGCCACUGGGCAAUCCUACUCAGGCGCAGCUAUCCAAGGUGCCGCACACCUUGUUCUACGAGAUGCCGCAGCA .(((((((.(((((....(((((((.(((((.......----)--)))))).(((((........)))))..))))))))))((((((.....)))))).............))))))). ( -47.10) >DroYak_CAF1 2085 114 + 1 AGCUGUGGAGGAUAACUCGCUGCGCAUCGGCUCCACAC----G--CUGGCCACUGGGCAAUCCCACGCAGGCGCAGCUAUCCAAAGUGCCACACACCUUGUUCUACGAGAUGCCGCAGCA .(((((((.(((((....(((((((..((((.......----)--)))((...((((....)))).))..))))))))))))...........((.((((.....)))).))))))))). ( -45.30) >DroMoj_CAF1 413358 114 + 1 UAGUGUAGUGAACAAUAAACUACGAAUUGGAGAAACAGAAAUGGAGCUGUCA--GAGAUAUC----ACGGGAUCAGCAAUCAAAAGUGCCCAGCACACUAUUCUAUGAUAUGCCACAGCA ...((((((.........))))))...........((....))..(((((..--(..(((((----(..((((.....)))....((((...))))......)..))))))..)))))). ( -22.70) >consensus UGCUGUGGAGGAUAACUCGCUGCGCAUCGGCUCCACAC____G__CUGGCCACUGGGCAAUCC_ACACAGGCGCAGCUAUCCAAGGUGCCGCACACCUUGUUCUACGAGAUGCCGCAGCA .(((((((..........((((((....((((...............))))...((....)).........))))))....(((((((.....)))))))............))))))). (-16.40 = -19.40 + 3.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:18 2006