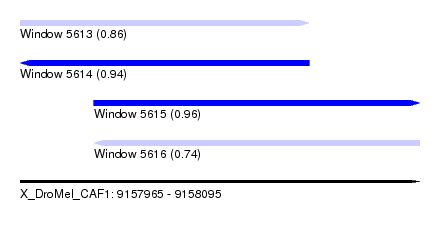
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,157,965 – 9,158,095 |
| Length | 130 |
| Max. P | 0.956664 |

| Location | 9,157,965 – 9,158,059 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.98 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -21.63 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.862783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
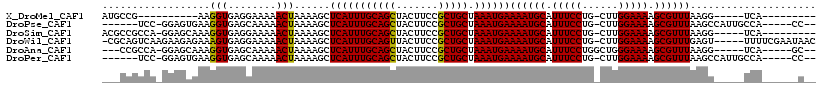
>X_DroMel_CAF1 9157965 94 + 22224390 ---------UGA-----CCUUAAACGCUUUUCCAAG-CAGGAAAUGCAUUUUCAUUUAGCAGCGGAAGUAGCUGCAAAUGAGCUUUUAGUUUUUCCUCACCUU----------CGGCAU ---------(((-----....((((((.(((((...-..))))).))...(((((((.(((((.......))))))))))))......))))....)))....----------...... ( -24.30) >DroPse_CAF1 10439 104 + 1 --GG-----UGGCAAUGGCUUAAACGCUUUUCCAAG-CAGGAAAUGCAUUUUCAUUUAGCAGCGGAAGUAGCUGCAAAUGAGCUUUUAGUUUUUGCUCACCUUCACUCC-GGA------ --((-----((((((..(((.(((.((.(((((...-..))))).))...(((((((.(((((.......)))))))))))).))).)))..)))).))))........-...------ ( -35.60) >DroSim_CAF1 1048 103 + 1 ---------UGA-----CCUUAAACGCUUUUCCAAG-CAGGAAAUGCAUUUUCAUUUAGCAGCGGAAGUAGCUGCAAAUGAGCUUUUAGUUUUUCCUCACCUUUGCUCC-UGGCGGCGU ---------...-----......(((((...(((((-(((((((.((...(((((((.(((((.......))))))))))))......)).))))).......))))..-))).))))) ( -27.91) >DroWil_CAF1 14648 112 + 1 GUUAUUCGAAAA-----ACUCAAACGCUUUUCCAAG-CAGGAAAUGCAUUUUCAUUUAGCAGCGGAAGUAACUGCAAAUGAGCUUUUAGUUUUUCCUCACUUUCUCUUCUUGACUGCG- .......(((((-----(((.(((.((.(((((...-..))))).))...(((((((.(((((....)...))))))))))).))).)))))))).......................- ( -25.20) >DroAna_CAF1 12832 103 + 1 --GC-----UGA-----CCUUAAACGCUUUUCCCAGCCAGGAAAUGCAUUUUCAUUUAGCAGCGGAAGUAGCUGCAAAUGAGCUUUUAGUUUUUGCUCACCUUUGCUCC-UGGCGG--- --..-----...-----...............((.(((((((...............(((.((....)).)))((((((((((...........)))))..))))))))-))))))--- ( -31.60) >DroPer_CAF1 9596 104 + 1 --GG-----UGGCAAUGGCUUAAACGCUUUUCCAAG-CAGGAAAUGCAUUUUCAUUUAGCAGCGGAAGUAGCUGCAAAUGAGCUUUUAGUUUUUGCUCACCUUCACUCC-GGA------ --((-----((((((..(((.(((.((.(((((...-..))))).))...(((((((.(((((.......)))))))))))).))).)))..)))).))))........-...------ ( -35.60) >consensus __G______UGA_____CCUUAAACGCUUUUCCAAG_CAGGAAAUGCAUUUUCAUUUAGCAGCGGAAGUAGCUGCAAAUGAGCUUUUAGUUUUUCCUCACCUUCACUCC_UGACGG___ .....................((((((.(((((......))))).))...(((((((.(((((.......))))))))))))......))))........................... (-21.63 = -21.80 + 0.17)



| Location | 9,157,965 – 9,158,059 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.98 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.30 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9157965 94 - 22224390 AUGCCG----------AAGGUGAGGAAAAACUAAAAGCUCAUUUGCAGCUACUUCCGCUGCUAAAUGAAAAUGCAUUUCCUG-CUUGGAAAAGCGUUUAAGG-----UCA--------- ..(((.----------..(((........)))......(((((((((((.......))))).))))))((((((.(((((..-...))))).))))))..))-----)..--------- ( -27.00) >DroPse_CAF1 10439 104 - 1 ------UCC-GGAGUGAAGGUGAGCAAAAACUAAAAGCUCAUUUGCAGCUACUUCCGCUGCUAAAUGAAAAUGCAUUUCCUG-CUUGGAAAAGCGUUUAAGCCAUUGCCA-----CC-- ------...-........((((.((((...........(((((((((((.......))))).))))))((((((.(((((..-...))))).))))))......))))))-----))-- ( -33.60) >DroSim_CAF1 1048 103 - 1 ACGCCGCCA-GGAGCAAAGGUGAGGAAAAACUAAAAGCUCAUUUGCAGCUACUUCCGCUGCUAAAUGAAAAUGCAUUUCCUG-CUUGGAAAAGCGUUUAAGG-----UCA--------- ..((((((.-........))))((......))....))(((((((((((.......))))).))))))((((((.(((((..-...))))).))))))....-----...--------- ( -28.60) >DroWil_CAF1 14648 112 - 1 -CGCAGUCAAGAAGAGAAAGUGAGGAAAAACUAAAAGCUCAUUUGCAGUUACUUCCGCUGCUAAAUGAAAAUGCAUUUCCUG-CUUGGAAAAGCGUUUGAGU-----UUUUCGAAUAAC -(((..((.......))..)))..((((((((......(((((((((((.......))))).))))))((((((.(((((..-...))))).)))))).)))-----)))))....... ( -31.90) >DroAna_CAF1 12832 103 - 1 ---CCGCCA-GGAGCAAAGGUGAGCAAAAACUAAAAGCUCAUUUGCAGCUACUUCCGCUGCUAAAUGAAAAUGCAUUUCCUGGCUGGGAAAAGCGUUUAAGG-----UCA-----GC-- ---((((((-((((((.((((((((...........))))))))(((((.......)))))..........)))...))))))).))...............-----...-----..-- ( -33.20) >DroPer_CAF1 9596 104 - 1 ------UCC-GGAGUGAAGGUGAGCAAAAACUAAAAGCUCAUUUGCAGCUACUUCCGCUGCUAAAUGAAAAUGCAUUUCCUG-CUUGGAAAAGCGUUUAAGCCAUUGCCA-----CC-- ------...-........((((.((((...........(((((((((((.......))))).))))))((((((.(((((..-...))))).))))))......))))))-----))-- ( -33.60) >consensus ___CCGUCA_GGAGCGAAGGUGAGCAAAAACUAAAAGCUCAUUUGCAGCUACUUCCGCUGCUAAAUGAAAAUGCAUUUCCUG_CUUGGAAAAGCGUUUAAGG_____UCA______C__ ..................(((........)))......(((((((((((.......))))).))))))((((((.(((((......))))).))))))..................... (-24.58 = -24.30 + -0.28)


| Location | 9,157,989 – 9,158,095 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -27.64 |
| Energy contribution | -28.03 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9157989 106 + 22224390 GGAAAUGCAUUUUCAUUUAGCAGCGGAAGUAGCUGCAAAUGAGCUUUUAGUUUUUCCUCACCUU---------CGGCAUUCUCGUGCGUCCGCAUUUCCGGUCCACCGUCCACCU---- ((((((((...(((((((.(((((.......)))))))))))).....................---------..((((....))))....))))))))((....))........---- ( -31.70) >DroSim_CAF1 1072 119 + 1 GGAAAUGCAUUUUCAUUUAGCAGCGGAAGUAGCUGCAAAUGAGCUUUUAGUUUUUCCUCACCUUUGCUCCUGGCGGCGUUGUCGUGCGUCCGCAUUUCCGGUCCACCGUCCACCUGGUC ((((((((....((((((.(((((.......)))))))))))(((...(((..............)))...)))(((((......))))).))))))))((....))(.((....)).) ( -37.04) >DroAna_CAF1 12859 101 + 1 GGAAAUGCAUUUUCAUUUAGCAGCGGAAGUAGCUGCAAAUGAGCUUUUAGUUUUUGCUCACCUUUGCUCCUGGCGG--------------CGCAUUUCCGGUCCGCCGUUAUCCU---- ((((((((....((((((.(((((.......)))))))))))(((.((((.....((........))..)))).))--------------)))))))))((....))........---- ( -34.70) >consensus GGAAAUGCAUUUUCAUUUAGCAGCGGAAGUAGCUGCAAAUGAGCUUUUAGUUUUUCCUCACCUUUGCUCCUGGCGGC_UU_UCGUGCGUCCGCAUUUCCGGUCCACCGUCCACCU____ ((((((((...(((((((.(((((.......))))))))))))................................(((......)))....))))))))((....))............ (-27.64 = -28.03 + 0.39)


| Location | 9,157,989 – 9,158,095 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -26.31 |
| Energy contribution | -26.87 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9157989 106 - 22224390 ----AGGUGGACGGUGGACCGGAAAUGCGGACGCACGAGAAUGCCG---------AAGGUGAGGAAAAACUAAAAGCUCAUUUGCAGCUACUUCCGCUGCUAAAUGAAAAUGCAUUUCC ----.(((.........)))(((((((((...((...((....((.---------.......)).....))....))(((((((((((.......))))).))))))...))))))))) ( -28.90) >DroSim_CAF1 1072 119 - 1 GACCAGGUGGACGGUGGACCGGAAAUGCGGACGCACGACAACGCCGCCAGGAGCAAAGGUGAGGAAAAACUAAAAGCUCAUUUGCAGCUACUUCCGCUGCUAAAUGAAAAUGCAUUUCC ..((.(((((.(((((..(((......))).))).))......))))).)).(((..(((........)))......(((((((((((.......))))).))))))...)))...... ( -33.80) >DroAna_CAF1 12859 101 - 1 ----AGGAUAACGGCGGACCGGAAAUGCG--------------CCGCCAGGAGCAAAGGUGAGCAAAAACUAAAAGCUCAUUUGCAGCUACUUCCGCUGCUAAAUGAAAAUGCAUUUCC ----........((....))(((((((((--------------(((((.........)))).)).............(((((((((((.......))))).))))))....)))))))) ( -32.10) >consensus ____AGGUGGACGGUGGACCGGAAAUGCGGACGCACGA_AA_GCCGCCAGGAGCAAAGGUGAGGAAAAACUAAAAGCUCAUUUGCAGCUACUUCCGCUGCUAAAUGAAAAUGCAUUUCC ............((....))(((((((((............((((............))))................(((((((((((.......))))).))))))...))))))))) (-26.31 = -26.87 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:15 2006