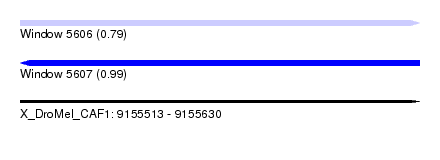
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,155,513 – 9,155,630 |
| Length | 117 |
| Max. P | 0.989751 |

| Location | 9,155,513 – 9,155,630 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -34.74 |
| Energy contribution | -34.82 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9155513 117 + 22224390 GUAUCGCCGGGUUAGCCGCGAAGGAGCCAAAUAGAGAUUGCCAAUUUGGCGGCCUCUCACCUCGCGAUAUUGGCCCAUUCACGAGCAUAAAUCAAAGCCAACA--AACCGGUUCCGGUU- .....(((((....((((.(((((.(((((...(((..((((.....))))..)))((.......))..))))))).))).)).)).........((((....--....))))))))).- ( -36.10) >DroSec_CAF1 8152 119 + 1 GAAUCGCCGGGUUAGCCGCGAAGGACCCAAAUAGAGAUUGCCAAUUUGGCGGCCUCUCACCUCGCGAUUUUGGCCCAUUCACGAGCAUAAAUCAAAGCCAACAAAAACCGGUUCCGGUU- (((((((.((((...((.....))))))....((((..((((.....))))..))))......)))))))((((......................)))).....(((((....)))))- ( -33.75) >DroSim_CAF1 9211 119 + 1 GAAUCGCCGGGUUAGCCGCGAAGGAGCCAAACAGAGAUUGCCAAUUUGGCGGCCUCUCACCUCGCGAUUUUGGCCCAUUCACGAGCAUAAAUCAAAGCCAACAAAAACCGGUUCCGGUU- .....(((((....((((.(((((.((((((..(((..((((.....))))..)))((.......)).)))))))).))).)).)).........((((..........))))))))).- ( -35.70) >DroEre_CAF1 8495 117 + 1 CAAUCGCCGGGUUAGCCGCGAAGGAGCCAAAUGGAGAUUGCCAAUUUGGCGGCCUCUCACCUCGCGAUUUUGGCCCAUUCCCGAGCGUGAACCAAAGCCACCA--AACCGGUUCCGGCU- .....((((((...((((....((.(((((((((......)).)))))))(((...((((((((.((...........)).)))).))))......))).)).--...)))))))))).- ( -42.10) >DroYak_CAF1 8658 118 + 1 CAAUCGCCGGGUUAGCCGCGAAGGAGCCAAAUAGAGAUUGCCAAUUUGGCGGCCUCUCACCUCGCGAUUUUGGCCCAUUCACGGGCAUAAAUCAAAGCCAACA--AACCGGUUCCGGUUG (((((((((((((.((.((((.((.(......((((..((((.....))))..))))).))))))(((((..((((......))))..)))))...)).))).--..))))...)))))) ( -41.10) >consensus GAAUCGCCGGGUUAGCCGCGAAGGAGCCAAAUAGAGAUUGCCAAUUUGGCGGCCUCUCACCUCGCGAUUUUGGCCCAUUCACGAGCAUAAAUCAAAGCCAACA__AACCGGUUCCGGUU_ .....(((((....((((.(((((.((((((.((((..((((.....))))..))))...........)))))))).))).)).)).........((((..........))))))))).. (-34.74 = -34.82 + 0.08)


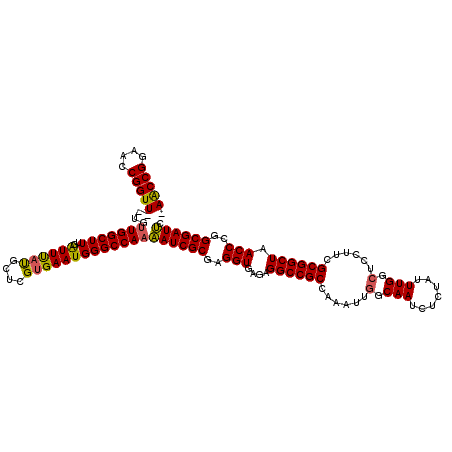
| Location | 9,155,513 – 9,155,630 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -45.40 |
| Consensus MFE | -39.48 |
| Energy contribution | -39.12 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9155513 117 - 22224390 -AACCGGAACCGGUU--UGUUGGCUUUGAUUUAUGCUCGUGAAUGGGCCAAUAUCGCGAGGUGAGAGGCCGCCAAAUUGGCAAUCUCUAUUUGGCUCCUUCGCGGCUAACCCGGCGAUAC -.((((....))))(--((((((.((........((.((((((.(((((((((((....))))(((((..(((.....)))..)))))..))))))).)))))))).)).)))))))... ( -44.40) >DroSec_CAF1 8152 119 - 1 -AACCGGAACCGGUUUUUGUUGGCUUUGAUUUAUGCUCGUGAAUGGGCCAAAAUCGCGAGGUGAGAGGCCGCCAAAUUGGCAAUCUCUAUUUGGGUCCUUCGCGGCUAACCCGGCGAUUC -(((((....))))).(((((((.((........((.((((((.(((((...(((....))).(((((..(((.....)))..))))).....))))))))))))).)).)))))))... ( -42.00) >DroSim_CAF1 9211 119 - 1 -AACCGGAACCGGUUUUUGUUGGCUUUGAUUUAUGCUCGUGAAUGGGCCAAAAUCGCGAGGUGAGAGGCCGCCAAAUUGGCAAUCUCUGUUUGGCUCCUUCGCGGCUAACCCGGCGAUUC -(((((....))))).(((((((.((........((.((((((.(((((((((((....))).(((((..(((.....)))..))))).)))))))).)))))))).)).)))))))... ( -45.00) >DroEre_CAF1 8495 117 - 1 -AGCCGGAACCGGUU--UGGUGGCUUUGGUUCACGCUCGGGAAUGGGCCAAAAUCGCGAGGUGAGAGGCCGCCAAAUUGGCAAUCUCCAUUUGGCUCCUUCGCGGCUAACCCGGCGAUUG -.(((((..((((((--((((((((((...((((.((((.((.(.......).)).))))))))))))))))))))))))..........((((((.......)))))).)))))..... ( -51.10) >DroYak_CAF1 8658 118 - 1 CAACCGGAACCGGUU--UGUUGGCUUUGAUUUAUGCCCGUGAAUGGGCCAAAAUCGCGAGGUGAGAGGCCGCCAAAUUGGCAAUCUCUAUUUGGCUCCUUCGCGGCUAACCCGGCGAUUG ..((((....))))(--((((((.((........(((.(((((.(((((((((((....))).(((((..(((.....)))..))))).)))))))).)))))))).)).)))))))... ( -44.50) >consensus _AACCGGAACCGGUU__UGUUGGCUUUGAUUUAUGCUCGUGAAUGGGCCAAAAUCGCGAGGUGAGAGGCCGCCAAAUUGGCAAUCUCUAUUUGGCUCCUUCGCGGCUAACCCGGCGAUUC .(((((....)))))....(((((((..((((((....)))))))))))))((((((..(((....((((((......(.(((.......))).)......)))))).)))..)))))). (-39.48 = -39.12 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:07 2006