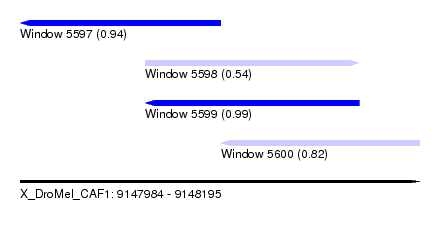
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,147,984 – 9,148,195 |
| Length | 211 |
| Max. P | 0.993631 |

| Location | 9,147,984 – 9,148,090 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 93.73 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -29.94 |
| Energy contribution | -29.98 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
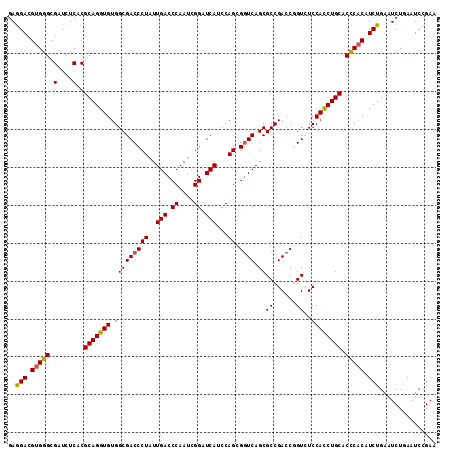
>X_DroMel_CAF1 9147984 106 - 22224390 AAUAGGGUCGCCACACCUGCGAGAGAUCGCCCACGUCCUCCUCCCUGGCGGAUUCCACUCCGCC---GGCACCACUGCCACCCACUUCGCCAGCGGAUUCCACCUGCUG---- ...((((((((.......))))(((..((....))..)))..))))((((((......))))))---((((....))))...........((((((.......))))))---- ( -35.80) >DroSec_CAF1 778 106 - 1 AAUAGGGUCGCCACACCUGCGAGAGAUCGCCCACGUCCUCCUCCCUGGCGGAUUCCACCCCGCC---GGCACCACUGCCACCCACUUCGCCAGCGGAUUCCACCUGCUG---- ...((((((((.......))))(((..((....))..)))..))))(((((........)))))---((((....))))...........((((((.......))))))---- ( -34.40) >DroSim_CAF1 390 106 - 1 AAUAGGGUCGCCACACCUGCGUGAGAUCGCCCACGUCUUCCUCCCUGGCGGAUUCCACCCCGCC---GGCACCACUGCCACCCACUUCGCCAGCGGAUUCCACCUGCUG---- ....((((.((.......(((((........)))))........(((((((........)))))---)).......)).)))).......((((((.......))))))---- ( -33.60) >DroEre_CAF1 777 110 - 1 AAUAGGGUCGCCACACCUGCGUGAGAUCGCCCACGUCCUCCUCCCUGGCGGAUUCCACCCCGCC---GGCACCACUGCCACCCACUUCGCCAGCGGACUCCACCUGCUGCUGC ....((((.((.......(((((........)))))........(((((((........)))))---)).......)).)))).....((.(((((.......)))))))... ( -34.40) >DroYak_CAF1 785 113 - 1 AAUAGGGUCGCCACACCUGCGUGAGAUCGCCCACGUCCUCCUCCCUGGCGGAUUCCACACCGCCACCCGCCACACUGCCACCCACUUCGCCAGCGGAUUCCACCUGCUGCUGC ....((((.((.......(((.(((..((....))..))).....((((((........))))))..)))......)).)))).....((.(((((.......)))))))... ( -32.72) >consensus AAUAGGGUCGCCACACCUGCGUGAGAUCGCCCACGUCCUCCUCCCUGGCGGAUUCCACCCCGCC___GGCACCACUGCCACCCACUUCGCCAGCGGAUUCCACCUGCUG____ ....((((.((......((((.(((..((....))..)))).....(((((........)))))....))).....)).)))).......((((((.......)))))).... (-29.94 = -29.98 + 0.04)


| Location | 9,148,050 – 9,148,163 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 93.81 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -32.38 |
| Energy contribution | -32.70 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9148050 113 + 22224390 GAGGACGUGGGCGAUCUCUCGCAGGUGUGGCGACCCUAUUGACCCAACCGGAUCAUUCAGCGAUCAGCGCCGACCGGUAUCCACCUGCACCCACAUCUGAAUCUGAAUCCGAA ..(((.(((((.((....))(((((((.(((((.....))).))..(((((.....((.(((.....))).)))))))...))))))).))))).((.......)).)))... ( -35.80) >DroSec_CAF1 844 113 + 1 GAGGACGUGGGCGAUCUCUCGCAGGUGUGGCGACCCUAUUGACCCAAUCGGAUCAUCCAGCGGUCAGCGCCGACCGGUAUCCACCUGCACCCACAUCUGAAUCUGAAUCCGAC ..(((.(((((.((....))(((((((.(((((((((..(((.((....)).)))...)).)))).))(((....)))..)))))))).))))).((.......)).)))... ( -39.70) >DroSim_CAF1 456 113 + 1 GAAGACGUGGGCGAUCUCACGCAGGUGUGGCGACCCUAUUGACCCAAUCGGAUCAUCCAGCGGUCAGCGCCGACCGGUCUCCACCUGCACCCACAUCUGAAUCUGAAUCCGAA ..(((.(((((.(....)..(((((((.((.((((....(((.((....)).)))......((((......))))))))))))))))).))))).)))............... ( -39.40) >DroEre_CAF1 847 113 + 1 GAGGACGUGGGCGAUCUCACGCAGGUGUGGCGACCCUAUUGACCCAAUCGGGUCAUCCAGAGGUCAGCGCCGACCAGUCUCCACCUGCACCCCCAUCUGAAUCUAAAUCCGAA ..(((.(((((....)))))(((((((.(((((((((..((((((....))))))...)).))))...)))((....))..)))))))...................)))... ( -42.30) >DroYak_CAF1 858 113 + 1 GAGGACGUGGGCGAUCUCACGCAGGUGUGGCGACCCUAUUGACCCAAUCGGAUCAACCAGAGGUCAGCGCCGACCAGUCUCCAUCUGCACUCACAUCUGAAUCUAAAUCCGAA ..(((((((((....))))))((((((((((((((((.((((.((....)).))))..)).)))).))((.((..........)).))...))))))))........)))... ( -35.50) >consensus GAGGACGUGGGCGAUCUCACGCAGGUGUGGCGACCCUAUUGACCCAAUCGGAUCAUCCAGCGGUCAGCGCCGACCGGUCUCCACCUGCACCCACAUCUGAAUCUGAAUCCGAA ..(((.(((((.(....)..(((((((.(((((((((..(((.((....)).)))...)).)))).))(((....)))..)))))))).))))).)))............... (-32.38 = -32.70 + 0.32)

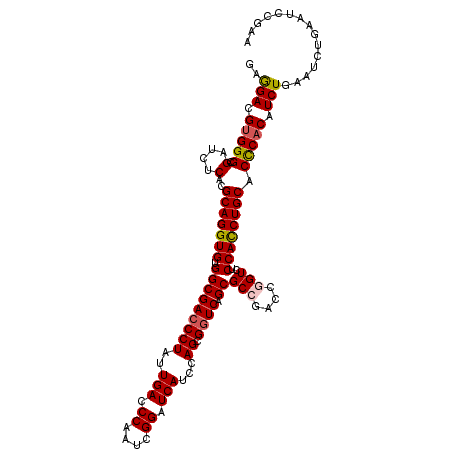
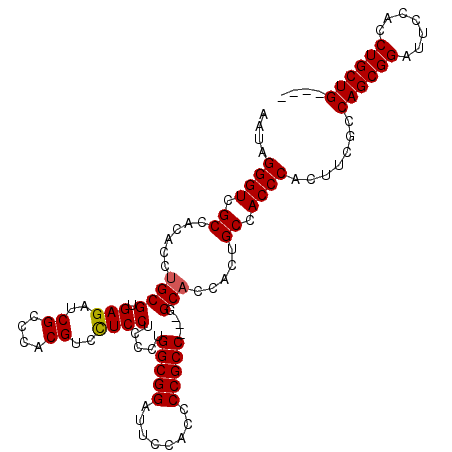
| Location | 9,148,050 – 9,148,163 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 93.81 |
| Mean single sequence MFE | -47.14 |
| Consensus MFE | -43.72 |
| Energy contribution | -44.16 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9148050 113 - 22224390 UUCGGAUUCAGAUUCAGAUGUGGGUGCAGGUGGAUACCGGUCGGCGCUGAUCGCUGAAUGAUCCGGUUGGGUCAAUAGGGUCGCCACACCUGCGAGAGAUCGCCCACGUCCUC ...((((....)))).((((((((((((((((...(((..((((((.....)))))).((((((....))))))....))).....)))))))((....)))))))))))... ( -46.70) >DroSec_CAF1 844 113 - 1 GUCGGAUUCAGAUUCAGAUGUGGGUGCAGGUGGAUACCGGUCGGCGCUGACCGCUGGAUGAUCCGAUUGGGUCAAUAGGGUCGCCACACCUGCGAGAGAUCGCCCACGUCCUC ...((((....)))).(((((((((((((((((((....)))(((...((((.(((..((((((....)))))).)))))))))).)))))))((....)))))))))))... ( -50.50) >DroSim_CAF1 456 113 - 1 UUCGGAUUCAGAUUCAGAUGUGGGUGCAGGUGGAGACCGGUCGGCGCUGACCGCUGGAUGAUCCGAUUGGGUCAAUAGGGUCGCCACACCUGCGUGAGAUCGCCCACGUCUUC ...((((....))))(((((((((((((((((..(((((((((....))))).(((..((((((....)))))).)))))))....)))))))..(....))))))))))).. ( -51.70) >DroEre_CAF1 847 113 - 1 UUCGGAUUUAGAUUCAGAUGGGGGUGCAGGUGGAGACUGGUCGGCGCUGACCUCUGGAUGACCCGAUUGGGUCAAUAGGGUCGCCACACCUGCGUGAGAUCGCCCACGUCCUC ...((((....)))).((((.(((((((((((..((....))(((...((((.(((..((((((....)))))).)))))))))).)))))))..(....))))).))))... ( -46.40) >DroYak_CAF1 858 113 - 1 UUCGGAUUUAGAUUCAGAUGUGAGUGCAGAUGGAGACUGGUCGGCGCUGACCUCUGGUUGAUCCGAUUGGGUCAAUAGGGUCGCCACACCUGCGUGAGAUCGCCCACGUCCUC ...((((.......((((.((.(((((.((((....)..))).))))).)).))))((((((((....)))))))).((((..((((......))).)...))))..)))).. ( -40.40) >consensus UUCGGAUUCAGAUUCAGAUGUGGGUGCAGGUGGAGACCGGUCGGCGCUGACCGCUGGAUGAUCCGAUUGGGUCAAUAGGGUCGCCACACCUGCGUGAGAUCGCCCACGUCCUC ...((((....)))).((((((((((((((((..(((((((((....))))).(((..((((((....)))))).)))))))....)))))))(......))))))))))... (-43.72 = -44.16 + 0.44)

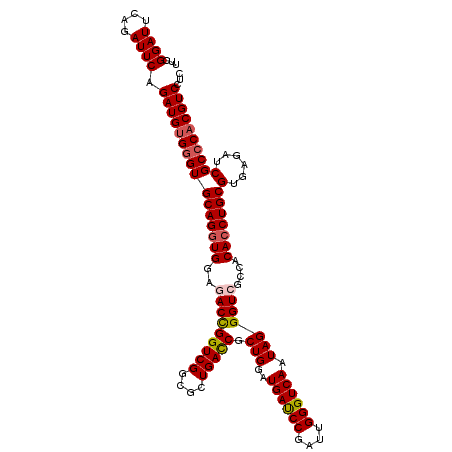
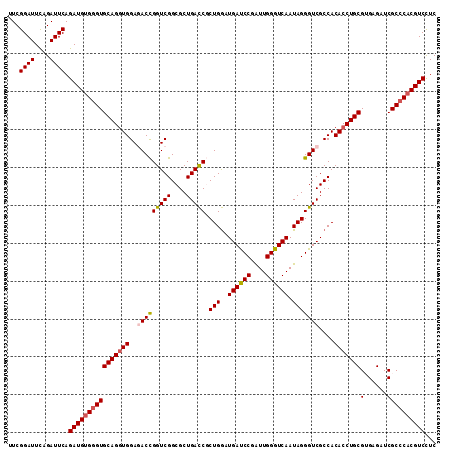
| Location | 9,148,090 – 9,148,195 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 86.08 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -28.50 |
| Energy contribution | -30.10 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9148090 105 - 22224390 GCAGCGGUGGGUGGCUGGG------AUUCAGAUUUGGAUUCGGAUUCAGAUUCAGAUGUGGGUGCAGGUGGAUACCGGUCGGCGCUGAUCGCUGAAUGAUCCGGUUGGGUC .(((((((.((((.((((.------(((((.((((((((..(....)..)))))))).))))).).(((....)))...))))))).)))))))...(((((....))))) ( -40.20) >DroSec_CAF1 884 105 - 1 GCAGCGGUGGGUGGCUGGG------AUUCGGAUUUGGAGUCGGAUUCAGAUUCAGAUGUGGGUGCAGGUGGAUACCGGUCGGCGCUGACCGCUGGAUGAUCCGAUUGGGUC .(((((((.(((((((((.------(((((((((((((......)))))))))...(((....)))...)))).))))))...))).)))))))...(((((....))))) ( -41.80) >DroSim_CAF1 496 105 - 1 GCAGCGGUGGGUGGCUGGG------AUUCGGAUUUGGAUUCGGAUUCAGAUUCAGAUGUGGGUGCAGGUGGAGACCGGUCGGCGCUGACCGCUGGAUGAUCCGAUUGGGUC .(((((((.(((((((((.------.(((((((((((((....))))))))))...(((....)))...)))..))))))...))).)))))))...(((((....))))) ( -43.20) >DroEre_CAF1 887 111 - 1 GCAGCGGUGGGUGGCUGGGAUUCGCAUUUGGAUUCGGAUUCGGAUUUAGAUUCAGAUGGGGGUGCAGGUGGAGACUGGUCGGCGCUGACCUCUGGAUGACCCGAUUGGGUC .(((.(((.((((.((((.((((.((((((((((..(((....)))..)))))))))).)))).)....(....)....))))))).))).)))...(((((....))))) ( -43.20) >DroYak_CAF1 898 102 - 1 GCAGCGGUG---------GAUUUGGAUUCGGAUACGGAUUCGGAUUUAGAUUCAGAUGUGAGUGCAGAUGGAGACUGGUCGGCGCUGACCUCUGGUUGAUCCGAUUGGGUC .((((..((---------((((((((((((....))))))))))))))....((((.((.(((((.((((....)..))).))))).)).))))))))((((....)))). ( -37.60) >consensus GCAGCGGUGGGUGGCUGGG______AUUCGGAUUUGGAUUCGGAUUCAGAUUCAGAUGUGGGUGCAGGUGGAGACCGGUCGGCGCUGACCGCUGGAUGAUCCGAUUGGGUC .(((((((.((((.(..........(((((.(((((((((........))))))))).)))))((.(((....))).)).).)))).)))))))...(((((....))))) (-28.50 = -30.10 + 1.60)

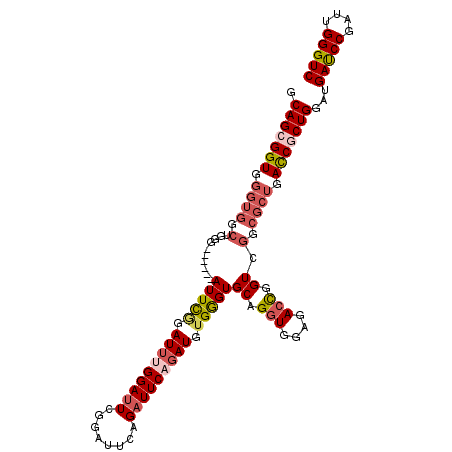
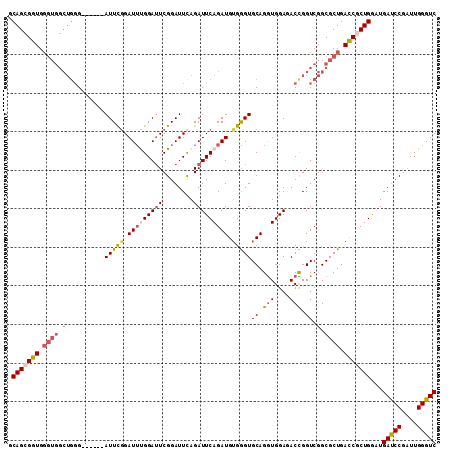
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:00 2006