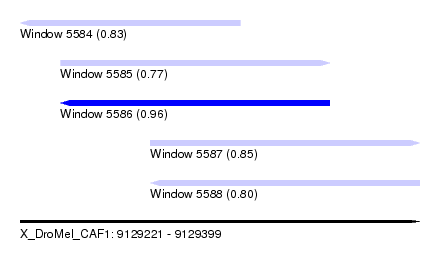
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,129,221 – 9,129,399 |
| Length | 178 |
| Max. P | 0.959334 |

| Location | 9,129,221 – 9,129,319 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.70 |
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -24.77 |
| Energy contribution | -25.15 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9129221 98 - 22224390 UCGCAUUUAAGCGCGCUCUCUUUAAUCAGCUGAGAGCGUGACGACUUAAGAGCGGCGCUCUUGUUCAGCCGGGAAAUUUCGAAAAUAUUUAGG--------------------AGCUG ((((.(((((((((((((((...........)))))))))....)))))).)))).(((((((......((((....))))........))))--------------------))).. ( -30.84) >DroSec_CAF1 5228 102 - 1 UCGCAGUUAAGCGCGCUCUCUUCAUUCAGCUGAGAGCGUGACGACUUAAGAGCGGCGCUCUUGUUGAGUCGGGAAAUUUCGAAAAUAUU----------------UACUUAAUAACUG ...((((((..(((((((((..(.....)..))))))))).((((((((((((...)))))...)))))))..................----------------.......)))))) ( -33.70) >DroSim_CAF1 3875 111 - 1 UCGC-------CGCGCUCUCUUCAUUCAGCUGAGAGCGUGACGACUUAAGAGCGGCGCUCUUGUUGAGUCGGGAAAUUUCGAAAAUAUUUAAGAGUUUACCUUUUUACCUAAUAACUG ....-------(((((((((..(.....)..))))))))).((((((((((((...)))))...)))))))((((((((..((.....))..)))))).))................. ( -31.50) >DroYak_CAF1 5175 115 - 1 UCGCAUACAA-CGCGCUCUCUUCAUUCAGCUGAGAGCGUGGCGACUUAAGAGCGGCGUUUUGGUCGAGCCGGGAAAUUUCGAAAAUAUUUAAGAGGCUUG--AGGAACUUAUUAUAUG ..((......-(((((((((..(.....)..)))))))))((.........)).))(((((..(((((((...((((.........))))....))))))--)))))).......... ( -32.10) >consensus UCGCAUUUAA_CGCGCUCUCUUCAUUCAGCUGAGAGCGUGACGACUUAAGAGCGGCGCUCUUGUUGAGCCGGGAAAUUUCGAAAAUAUUUAAG____________UACUUAAUAACUG (((........(((((((((...........))))))))).((((((((((((...)))))))...)))))........))).................................... (-24.77 = -25.15 + 0.38)



| Location | 9,129,239 – 9,129,359 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -42.08 |
| Consensus MFE | -31.75 |
| Energy contribution | -33.62 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9129239 120 + 22224390 GAAAUUUCCCGGCUGAACAAGAGCGCCGCUCUUAAGUCGUCACGCUCUCAGCUGAUUAAAGAGAGCGCGCUUAAAUGCGAUUCCAAUUGGAAGUGGUUGCAGCCGGUGGUGGAUGAAAAU ......((((((((((((..((((((.((((((...(.((((.((.....)))))).)..)))))))))))).....((.((((....)))).))))).))))))).))........... ( -43.10) >DroSec_CAF1 5250 120 + 1 GAAAUUUCCCGACUCAACAAGAGCGCCGCUCUUAAGUCGUCACGCUCUCAGCUGAAUGAAGAGAGCGCGCUUAACUGCGAUUCCAAUUGGAAGUGGUUGCAGCCGGCGGUGGAUGAAAAU ...(((..((((((....((((((...)))))).))))(((.(((((((..(.....)..))))))).(((((((..(...(((....))).)..)))).))).)))))..)))...... ( -41.00) >DroSim_CAF1 3913 113 + 1 GAAAUUUCCCGACUCAACAAGAGCGCCGCUCUUAAGUCGUCACGCUCUCAGCUGAAUGAAGAGAGCGCG-------GCGAUUCCAAUUGGAAGUGGUUGCAGCCGGCGGUGGAUGAAAAU ....((((....(((.....)))(((((((......(((((.(((((((..(.....)..))))))).)-------))))((((....))))..(((....))))))))))...)))).. ( -40.00) >DroYak_CAF1 5211 119 + 1 GAAAUUUCCCGGCUCGACCAAAACGCCGCUCUUAAGUCGCCACGCUCUCAGCUGAAUGAAGAGAGCGCG-UUGUAUGCGAUUCCAAUUGGAAGUGGUUGCAGUCGGCGGUGGUUGAAAAU .............(((((((...(((((((.....((.(((((((((((..(.....)..))))))(((-(...))))..((((....))))))))).)))).))))).))))))).... ( -44.20) >consensus GAAAUUUCCCGACUCAACAAGAGCGCCGCUCUUAAGUCGUCACGCUCUCAGCUGAAUGAAGAGAGCGCG_UUAAAUGCGAUUCCAAUUGGAAGUGGUUGCAGCCGGCGGUGGAUGAAAAU ......((((((((((((..((((((.((((((...((((((.((.....))))).))).)))))))))))).....((.((((....)))).)))))).)))))).))........... (-31.75 = -33.62 + 1.88)


| Location | 9,129,239 – 9,129,359 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -40.08 |
| Consensus MFE | -33.69 |
| Energy contribution | -33.50 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9129239 120 - 22224390 AUUUUCAUCCACCACCGGCUGCAACCACUUCCAAUUGGAAUCGCAUUUAAGCGCGCUCUCUUUAAUCAGCUGAGAGCGUGACGACUUAAGAGCGGCGCUCUUGUUCAGCCGGGAAAUUUC ..............(((((((.(((...((((....))))((((.(((((((((((((((...........)))))))))....)))))).)))).......))))))))))........ ( -42.10) >DroSec_CAF1 5250 120 - 1 AUUUUCAUCCACCGCCGGCUGCAACCACUUCCAAUUGGAAUCGCAGUUAAGCGCGCUCUCUUCAUUCAGCUGAGAGCGUGACGACUUAAGAGCGGCGCUCUUGUUGAGUCGGGAAAUUUC .......(((......((((((......((((....))))..))))))...(((((((((..(.....)..))))))))).((((((((((((...)))))...))))))))))...... ( -40.80) >DroSim_CAF1 3913 113 - 1 AUUUUCAUCCACCGCCGGCUGCAACCACUUCCAAUUGGAAUCGC-------CGCGCUCUCUUCAUUCAGCUGAGAGCGUGACGACUUAAGAGCGGCGCUCUUGUUGAGUCGGGAAAUUUC ..............((((((.((((...((((....))))(((.-------(((((((((..(.....)..))))))))).)))...((((((...))))))))))))))))........ ( -40.50) >DroYak_CAF1 5211 119 - 1 AUUUUCAACCACCGCCGACUGCAACCACUUCCAAUUGGAAUCGCAUACAA-CGCGCUCUCUUCAUUCAGCUGAGAGCGUGGCGACUUAAGAGCGGCGUUUUGGUCGAGCCGGGAAAUUUC ...(((.((((.(((((.((........((((....))))((((......-(((((((((..(.....)..)))))))))))))......)))))))...)))).)))............ ( -36.90) >consensus AUUUUCAUCCACCGCCGGCUGCAACCACUUCCAAUUGGAAUCGCAUUUAA_CGCGCUCUCUUCAUUCAGCUGAGAGCGUGACGACUUAAGAGCGGCGCUCUUGUUGAGCCGGGAAAUUUC ..............((((((........((((....))))...........(((((((((...........))))))))).((((...(((((...))))).))))))))))........ (-33.69 = -33.50 + -0.19)



| Location | 9,129,279 – 9,129,399 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.60 |
| Mean single sequence MFE | -43.95 |
| Consensus MFE | -38.70 |
| Energy contribution | -39.08 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9129279 120 + 22224390 CACGCUCUCAGCUGAUUAAAGAGAGCGCGCUUAAAUGCGAUUCCAAUUGGAAGUGGUUGCAGCCGGUGGUGGAUGAAAAUGUGUCCACCAAUGGCGCGCCUCAAAUGCCAUGUCCACCUG ..(((((((...........)))))))(((......)))........((((.((((((((.((((.((((((((........)))))))).)))))))........))))).)))).... ( -45.90) >DroSec_CAF1 5290 120 + 1 CACGCUCUCAGCUGAAUGAAGAGAGCGCGCUUAACUGCGAUUCCAAUUGGAAGUGGUUGCAGCCGGCGGUGGAUGAAAAUGUGUCCACCAAUGGCGCGCCUCAAAUGCCAUGUCCACCUG ..(((((((..(.....)..)))))))(((......)))........((((.((((((((.((((..(((((((........)))))))..)))))))........))))).)))).... ( -45.50) >DroSim_CAF1 3953 113 + 1 CACGCUCUCAGCUGAAUGAAGAGAGCGCG-------GCGAUUCCAAUUGGAAGUGGUUGCAGCCGGCGGUGGAUGAAAAUGUGUCCACCAAUGGCGCGCCUCAAAUGCCAUGUCCACCUG ..(((((((..(.....)..))))))).(-------(((.((((....))))(.(((.((.((((..(((((((........)))))))..))))))))).)...))))........... ( -45.60) >DroYak_CAF1 5251 119 + 1 CACGCUCUCAGCUGAAUGAAGAGAGCGCG-UUGUAUGCGAUUCCAAUUGGAAGUGGUUGCAGUCGGCGGUGGUUGAAAAUGUGUCCACCAAUGGCGCGCCUCAAAUGCCGUGUCCACCUG ..(((((((..(.....)..))))))).(-(((..(((((((((....)))....))))))..))))(((((............)))))..(((((((.(......).)))).))).... ( -38.80) >consensus CACGCUCUCAGCUGAAUGAAGAGAGCGCG_UUAAAUGCGAUUCCAAUUGGAAGUGGUUGCAGCCGGCGGUGGAUGAAAAUGUGUCCACCAAUGGCGCGCCUCAAAUGCCAUGUCCACCUG ..(((((((...........)))))))(((......)))........((((..(((((((.((((..(((((((........)))))))..)))))))........))))..)))).... (-38.70 = -39.08 + 0.37)


| Location | 9,129,279 – 9,129,399 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.60 |
| Mean single sequence MFE | -41.08 |
| Consensus MFE | -38.29 |
| Energy contribution | -38.35 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9129279 120 - 22224390 CAGGUGGACAUGGCAUUUGAGGCGCGCCAUUGGUGGACACAUUUUCAUCCACCACCGGCUGCAACCACUUCCAAUUGGAAUCGCAUUUAAGCGCGCUCUCUUUAAUCAGCUGAGAGCGUG ..(((((.....((.......))(((((..(((((((..........)))))))..))).))..)))))(((....)))...((......))((((((((...........)))))))). ( -41.90) >DroSec_CAF1 5290 120 - 1 CAGGUGGACAUGGCAUUUGAGGCGCGCCAUUGGUGGACACAUUUUCAUCCACCGCCGGCUGCAACCACUUCCAAUUGGAAUCGCAGUUAAGCGCGCUCUCUUCAUUCAGCUGAGAGCGUG ..(((((.....((.......))(((((..(((((((..........)))))))..))).))..)))))(((....)))...((......))((((((((..(.....)..)))))))). ( -41.40) >DroSim_CAF1 3953 113 - 1 CAGGUGGACAUGGCAUUUGAGGCGCGCCAUUGGUGGACACAUUUUCAUCCACCGCCGGCUGCAACCACUUCCAAUUGGAAUCGC-------CGCGCUCUCUUCAUUCAGCUGAGAGCGUG ((((((.......)))))).((((((((..(((((((..........)))))))..))).))......((((....))))..))-------)((((((((..(.....)..)))))))). ( -42.60) >DroYak_CAF1 5251 119 - 1 CAGGUGGACACGGCAUUUGAGGCGCGCCAUUGGUGGACACAUUUUCAACCACCGCCGACUGCAACCACUUCCAAUUGGAAUCGCAUACAA-CGCGCUCUCUUCAUUCAGCUGAGAGCGUG ..(((((.((((((......((....))...(((((............)))))))))..))...)))))(((....)))...........-(((((((((..(.....)..))))))))) ( -38.40) >consensus CAGGUGGACAUGGCAUUUGAGGCGCGCCAUUGGUGGACACAUUUUCAUCCACCGCCGGCUGCAACCACUUCCAAUUGGAAUCGCAUUUAA_CGCGCUCUCUUCAUUCAGCUGAGAGCGUG ..(((((.....((.......))(((((..((((((............))))))..))).))..)))))(((....)))............(((((((((...........))))))))) (-38.29 = -38.35 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:49 2006