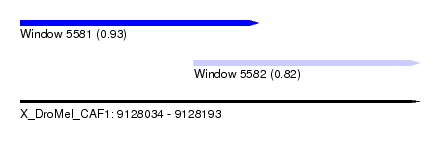
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,128,034 – 9,128,193 |
| Length | 159 |
| Max. P | 0.928469 |

| Location | 9,128,034 – 9,128,129 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 94.53 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -35.90 |
| Energy contribution | -36.70 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
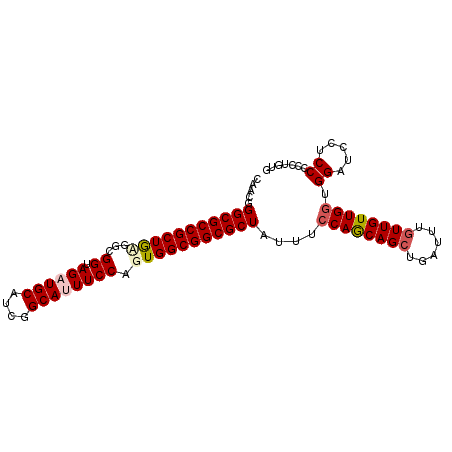
>X_DroMel_CAF1 9128034 95 + 22224390 CAUUGGGCGCCGCUGUGGCGGUAGAUGCAUCGGCAUUUCCAGUGGCGGCGCUAUUUCCAGCAGCUGAUUUGUUGUUGGCGGAUCCUCCCCCUGUG (((.(((((((((..(...((.((((((....)))))))).)..))))))).....((((((((......)))))))).((.....)).)).))) ( -40.50) >DroSec_CAF1 3859 95 + 1 CAACGGGCGCCGCUGCGGCGGUAGAUGCAUCGGCAUUUCCAGUGGCGGCGCUAUUUACAACAGCUGAUUUAUUGUUGGUGGAUCCUCCCCCUGUG ..(((((((((((..(...((.((((((....)))))))).)..))))))).(((((((((((........))))).)))))).......)))). ( -36.20) >DroSim_CAF1 2486 95 + 1 CAACGGGCGCCGCUACGGCGGUAGAUGCAUCGGCAUUUCCAGUGGCGGCGCUAUUUCCAGCAGCUGAUUUGUUGUUGGUGGAUCCUCCCCCUGUG ..((((((((((((((...((.((((((....)))))))).)))))))))).....((((((((......)))))))).((.....))..)))). ( -43.80) >DroEre_CAF1 3842 95 + 1 CAACGGGCGCCGCUGAGGCGGCAGCUGCAUCGGCAUUUCCAGUGGCGGCGCUAUUUCCAGCAGCUGAUUUGUUGUUGGUGGAUCCUCCCCCUGCG .....((((((((....))))).)))(((..((....(((((((((...)))))..((((((((......)))))))))))).....))..))). ( -35.90) >DroYak_CAF1 3933 95 + 1 CAACGGGCGCCGCUGAGGCGGCAGCUGCAUCGGCAUUUCCAGUGGCGGCGCUAUUUCCAGCAGCUGAUUUGUUGUUGGUGGAUCCUCCCCCUGUG ..(((((.(((((((.(((....)))((....)).....)))))))((..(((...((((((((......)))))))))))..))....))))). ( -36.30) >consensus CAACGGGCGCCGCUGAGGCGGUAGAUGCAUCGGCAUUUCCAGUGGCGGCGCUAUUUCCAGCAGCUGAUUUGUUGUUGGUGGAUCCUCCCCCUGUG .....(((((((((((...((.((((((....)))))))).)))))))))))....((((((((......)))))))).((.....))....... (-35.90 = -36.70 + 0.80)

| Location | 9,128,103 – 9,128,193 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -28.01 |
| Energy contribution | -27.95 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9128103 90 + 22224390 UGUUGUUGGCGGAUCCUCCCCCUGUGGCAGCACCACUACC------ACCGCCGCCAACCCCUCGGCCGUCCAUCUUCUUGGUGGAAUUGCGCGUUC ....((((((((...........((((.....))))....------....))))))))....((((..((((((.....))))))...)).))... ( -29.81) >DroSec_CAF1 3928 90 + 1 UAUUGUUGGUGGAUCCUCCCCCUGUGGCAGCACCACCACC------GCCGCCGCCAACCCCUCGCCCAUCCAUCUUCUUGGUGGAAUUGCGCGUUC ....((((((((...........((((.....))))....------....))))))))....(((.((((((((.....))))))..)).)))... ( -27.41) >DroSim_CAF1 2555 90 + 1 UGUUGUUGGUGGAUCCUCCCCCUGUGGCAGCACCACCACC------GCCGCCGCCAACCCCUCGCCCGUCCAUCUUCUUGGUGGAAUUGCGCGUUC ....((((((((...........((((.....))))....------....))))))))..(.(((...((((((.....))))))...))).)... ( -28.21) >DroYak_CAF1 4002 96 + 1 UGUUGUUGGUGGAUCCUCCCCCUGUGGCAGCGCCACCCCCGCCGCCACCGCCAGCAGCCCCUCGCCCAUCCAUCUUCUUGGUGGAAUUGCGCGUUC .(((((((((((.....))....(((((.(((.......))).))))).)))))))))....(((.((((((((.....))))))..)).)))... ( -36.90) >consensus UGUUGUUGGUGGAUCCUCCCCCUGUGGCAGCACCACCACC______ACCGCCGCCAACCCCUCGCCCAUCCAUCUUCUUGGUGGAAUUGCGCGUUC .((((.((((((...........((((.....))))...........)))))).))))..(.(((...((((((.....))))))...))).)... (-28.01 = -27.95 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:44 2006