
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,103,230 – 9,103,344 |
| Length | 114 |
| Max. P | 0.670315 |

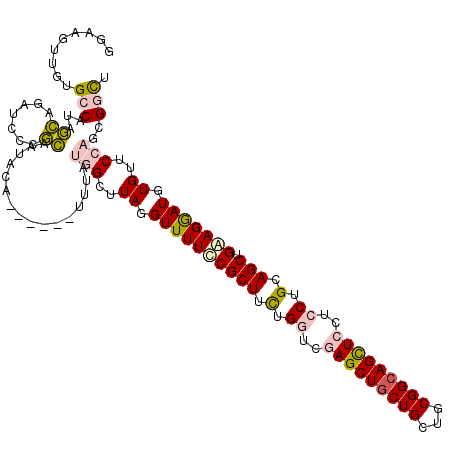
| Location | 9,103,230 – 9,103,344 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -42.11 |
| Consensus MFE | -27.31 |
| Energy contribution | -27.57 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9103230 114 - 22224390 GGAAGUUGUGCCAUAGCAGAUCCCGCAAUACA------UUUGUGGCUUAGGUUUUCCGCUUCUGGUCGAGCUGCUGCUGCGGCAGCUCCUCCUGCAGCUGAAGGAUGUGUUCCAGCGGCU (((((.(.(((....))).)..((((((....------.)))))).......)))))((....((..(((((((((...)))))))))..)).))(((((..(((.....)))..))))) ( -42.40) >DroPse_CAF1 10375 112 - 1 --------UGACAUCAUCGAUAUCAUUAUGCAGGCUAUGUUACUGCUUAGGUUUUGCGCUUCUGCUCAAGCUGCUGCUGCGGCAGUUCCUCCUGGAGCUGCAGAAUGUGCUCCAGCGGCU --------((((((..((..(((....)))..))..))))))(((((..((....((((((((((....(((((....)))))((((((....)))))))))))).)))).))))))).. ( -40.20) >DroSim_CAF1 8909 114 - 1 GGAAGUUGUGCCAUAGCAGAUCCCGCAAUACA------UUUGUGGCUUAGGUUUUCCGCUUCUGGUCGAGCUGCUGCUGCGGAAGCUCCUCCUGCAGCUGAAGGAUGUGUUCCAGCGGCU .....(((.((((.(((.....((((((....------.))))))....((....)))))..)))))))((((((((((((((......))).)))))....(((.....))))))))). ( -40.40) >DroEre_CAF1 9964 114 - 1 GGAAGUUGUGCCAUAGCAGAUCCCGCAAUACA------UUUCUGGCUUAGGUUUUCCGCUUUUGGUCGAGCUGCUGCUGCGGCAGCUCCUCCUGCAGCUGGAGGAUGUGUUCCAGGGGUU ........(((....)))((((((..((((((------((((..(((..((....))((....((..(((((((((...)))))))))..)).)))))..)).))))))))...)))))) ( -45.60) >DroYak_CAF1 10371 114 - 1 GGAAGUUGUGCCAUAGCAGAUCCCGCAAUACA------UUUCUGGCUUAGGUUUUCCGCUUCUGGUCGAGCUGCUGCUGCGGCAGCUCCUCCUGCAGCUGGAGGAUGUGUUCCAGCGGCU ((((((.(.(((..((((((............------..))).)))..)))...).))))))((..(((((((((...)))))))))..)).((.(((((((......))))))).)). ( -44.14) >DroPer_CAF1 11218 112 - 1 --------UGACAUCAUCGAUAUCAUUAUGCAGGCUAUGCUACUGCUUAGGUUUUGCGCUUCUGCUCAAGCUGCUGCUGCGGCAGUUCCUCCUGGAGCUGCAGAAUGUGUUCCAGCGGUU --------(((.(((...))).)))....(((.....))).((((((..((....((((((((((....(((((....)))))((((((....)))))))))))).)))).)))))))). ( -39.90) >consensus GGAAGUUGUGCCAUAGCAGAUCCCGCAAUACA______UUUAUGGCUUAGGUUUUCCGCUUCUGGUCGAGCUGCUGCUGCGGCAGCUCCUCCUGCAGCUGAAGGAUGUGUUCCAGCGGCU .........(((...((.......))................(((..((.((((((((((.(.((..(((((((((...)))))))))..)).).))).))))))).))..)))..))). (-27.31 = -27.57 + 0.26)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:27 2006