
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,096,333 – 9,096,438 |
| Length | 105 |
| Max. P | 0.709982 |

| Location | 9,096,333 – 9,096,438 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.31 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -21.90 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
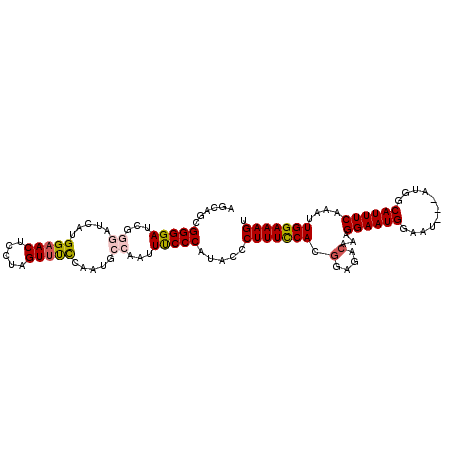
>X_DroMel_CAF1 9096333 105 + 22224390 AGCAGCGGGGAUCGGGAUCAUGGAACUCCUAGUUUCCAAUGCCAAUUCCCCAUACCCUUUCCACGGAGACAAAGGAAUGGAAU---AUGGCAUUUCAAAUUGGAAAGU ......(((((...((....(((((........)))))...))...))))).....(((((((.(....)...((((((....---....))))))....))))))). ( -32.30) >DroSec_CAF1 3421 105 + 1 AGCAGCGGGGAUCGGGAUCAUGGAACCUCUAGUUUCCAAUGCCAAUUUCCCAUACCCUUUCCACGGAGACAAAGGAAUGGAAU---AUGGCAUUUCAAAUUGGAAAGU ......(((((...((....(((((.(....).)))))...))...))))).....(((((((.(....)...((((((....---....))))))....))))))). ( -29.80) >DroSim_CAF1 2068 105 + 1 AGCAGCGGGGAUCGGGAUCAUGGAACCUCUAGUUUCCAAUGCCAAUUUCCCAUGCCCUUUCCACGGAGACAAAGGAAUGGAAU---AUGGCAUUUCAAAUUGGAAAGU .(((..(((((...((....(((((.(....).)))))...))...))))).))).(((((((.(....)...((((((....---....))))))....))))))). ( -32.90) >DroEre_CAF1 3361 102 + 1 AGCAGUGGGGA------UCGUGGAACUCCUAGUUCUCAAUUCCCAUUUCCCAUACCCUUUUCAUGAAGAAAAAGGAAUGGAAUGGAAUGGCAUUUCGAAUUGGAAAGU .((((((((((------(.(.(((((.....)))))).)))))))))..((((.((.(((((.....))))).)).)))).........)).((((......)))).. ( -28.20) >DroYak_CAF1 3528 99 + 1 AUUAGUGGGGA------UCAUGGAACUCCAAGUGCCCAAUGCCAAUUUCCCAUACCCUUUCCACGAAGACAAAGGAAUGGAAU---AUGGCAUUUCAAAUUGGAAAGU .....((((.(------(..(((....))).)).))))...(((((((.(((((.((.((((...........)))).))..)---))))......)))))))..... ( -23.40) >consensus AGCAGCGGGGAUCGGGAUCAUGGAACUCCUAGUUUCCAAUGCCAAUUUCCCAUACCCUUUCCACGGAGACAAAGGAAUGGAAU___AUGGCAUUUCAAAUUGGAAAGU ......(((((...((.....(((((.....))))).....))...))))).....(((((((.(....)...((((((...........))))))....))))))). (-21.90 = -22.38 + 0.48)

| Location | 9,096,333 – 9,096,438 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.31 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -21.76 |
| Energy contribution | -21.16 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9096333 105 - 22224390 ACUUUCCAAUUUGAAAUGCCAU---AUUCCAUUCCUUUGUCUCCGUGGAAAGGGUAUGGGGAAUUGGCAUUGGAAACUAGGAGUUCCAUGAUCCCGAUCCCCGCUGCU .(((((((....((((((....---....))))......))....)))))))((((((((((.((((...(((((........))))).....)))))))))).)))) ( -28.70) >DroSec_CAF1 3421 105 - 1 ACUUUCCAAUUUGAAAUGCCAU---AUUCCAUUCCUUUGUCUCCGUGGAAAGGGUAUGGGAAAUUGGCAUUGGAAACUAGAGGUUCCAUGAUCCCGAUCCCCGCUGCU ..((((((.......((((((.---.((((((.(((((..(.....)..))))).))))))...)))))))))))).(((.((......(((....))).)).))).. ( -25.51) >DroSim_CAF1 2068 105 - 1 ACUUUCCAAUUUGAAAUGCCAU---AUUCCAUUCCUUUGUCUCCGUGGAAAGGGCAUGGGAAAUUGGCAUUGGAAACUAGAGGUUCCAUGAUCCCGAUCCCCGCUGCU .(((((((....((((((....---....))))......))....)))))))((((((((..(((((...(((((.(....).))))).....))))).)))).)))) ( -26.00) >DroEre_CAF1 3361 102 - 1 ACUUUCCAAUUCGAAAUGCCAUUCCAUUCCAUUCCUUUUUCUUCAUGAAAAGGGUAUGGGAAAUGGGAAUUGAGAACUAGGAGUUCCACGA------UCCCCACUGCU .................((.......((((((.((((((((.....)))))))).))))))..((((.((((.((((.....))))..)))------).))))..)). ( -28.30) >DroYak_CAF1 3528 99 - 1 ACUUUCCAAUUUGAAAUGCCAU---AUUCCAUUCCUUUGUCUUCGUGGAAAGGGUAUGGGAAAUUGGCAUUGGGCACUUGGAGUUCCAUGA------UCCCCACUAAU ..............(((((((.---.((((((.(((((.(((....)))))))).))))))...)))))))(((....(((....)))...------..)))...... ( -29.60) >consensus ACUUUCCAAUUUGAAAUGCCAU___AUUCCAUUCCUUUGUCUCCGUGGAAAGGGUAUGGGAAAUUGGCAUUGGAAACUAGGAGUUCCAUGAUCCCGAUCCCCGCUGCU (((((....(((..(((((((.....((((((.(((((.((.....)).))))).))))))...)))))))..)))...)))))........................ (-21.76 = -21.16 + -0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:24 2006