| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,084,548 – 9,084,654 |
| Length | 106 |
| Max. P | 0.873082 |

| Location | 9,084,548 – 9,084,654 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.12 |
| Mean single sequence MFE | -43.30 |
| Consensus MFE | -22.16 |
| Energy contribution | -22.48 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
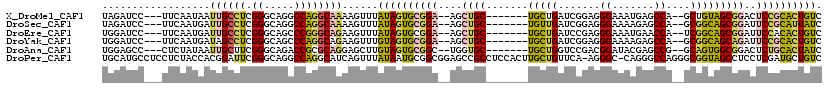
>X_DroMel_CAF1 9084548 106 + 22224390 UAGAUCC---UUCAAUAAUUGCCUCGGGCAGGCCAGGCAAAAGUUUAUAGUGCGGA--AGCUGC-------UGCUGAUCGGAGGGAAAUGAGCCA--GCUGUAGCGGACUCCGCACUGUC .......---........((((((.((.....))))))))......((((((((((--.(((((-------.((((.(((........)))..))--)).)))))....)))))))))). ( -40.90) >DroSec_CAF1 11698 106 + 1 UAGAUCC---UUCAAUGAUUGCCUCGGGCAGGCCAGGCAAAAGUUUAUAGUGCGGA--AGCUGC-------UGUUGAUCGGAGGGAAAAGAGCCA--GCGGCAGCGGAUUCCGCAUUAUC .......---........((((((.((.....))))))))......((((((((((--((((((-------(((((.((..........))..))--))))))))...))))))))))). ( -41.20) >DroEre_CAF1 15067 106 + 1 UGGAUCC---UUCAAUGAUUGCCUCGGGCAGCCCGGGCAGAAGUUUAUAGUGCGGA--AGCUGC-------UGCUGAUCCGAGGGAAAUGAACCA--UCGGCAGCGGAUUCCACACUGUC ......(---(((......((((.((((...))))))))))))...((((((.(((--(.((((-------(((((((....((........)))--)))))))))).)))).)))))). ( -45.00) >DroYak_CAF1 11297 106 + 1 UGGAUCC---UUCAAUGAUAGCCUCGGGCAGCCCAGGCAGAAGUUUGUAGUGCGGA--AGCUGC-------UGCUGAUCGGAGGGAAAAGAGCCA--GCGGCAGCAGAUUCCGCACUGUC .......---..........((((.(((...))))))).........(((((((((--((((((-------(((((.((..........))..))--))))))))...)))))))))).. ( -45.10) >DroAna_CAF1 12631 106 + 1 UGGAGCC---CUCUAUAAUUGCUUCGGGCAGACCGCGCAGGAGCUUGUAGUGCGGC--UGGUGC-------UGCUGGUCCGACGGAUACGAGCCG--GCAGUGGCGGACUCUGCACUAUC ..(((((---((........((..(((.....))).))))).))))((((((((((--((.(((-------((((((..((.......))..)))--)))))).)))...))))))))). ( -44.10) >DroPer_CAF1 13024 118 + 1 UGCAUGCCUCCUCUACCACGGAUUCGGGCAGGCCAGGCAUCAGUUUAUAAUGCGGCGGAGCCGCCUCCACUUGCUGUUCA-AGGGC-CAGGGCCAGGGCGGUAGCCUCCUCGAUGCUGUC .((.(((((..(((.....)))...))))).))..((((((.((.......))(((...(((((((...((((.....))-))(((-....))).))))))).))).....))))))... ( -43.50) >consensus UGGAUCC___UUCAAUGAUUGCCUCGGGCAGGCCAGGCAGAAGUUUAUAGUGCGGA__AGCUGC_______UGCUGAUCGGAGGGAAAAGAGCCA__GCGGCAGCGGACUCCGCACUGUC ..................((((((.((.....))))))))......((((((((((....((((.......(((((.......((.......))....)))))))))..)))))))))). (-22.16 = -22.48 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:21 2006