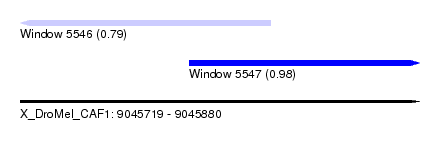
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 9,045,719 – 9,045,880 |
| Length | 161 |
| Max. P | 0.976082 |

| Location | 9,045,719 – 9,045,820 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.50 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -12.15 |
| Energy contribution | -16.40 |
| Covariance contribution | 4.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9045719 101 - 22224390 UCCUCUUCGU---GGGGUGGCACCAUUUAUUUUUG----GGGUUCUGAAACUUUUGGUGGAAAAACAGCAGAUACAGAAACGUAUAGAAAACCGGGUGGUGCCAAUAU .((((.....---))))((((((((((......((----(..(((((.....((((.((......)).))))(((......))))))))..))))))))))))).... ( -32.10) >DroSim_CAF1 3599 99 - 1 UCCUCUUCGU---GGGGUGGCACCAUUUAUUUUG------GGGUCUGAAACUGUUGGAGGAAAAACAGCAGAAACAGAAGCGUAUAAAAAACCGGGUGGUGCCAAUAU .((((.....---))))(((((((((((.(((((------...((((...((((((.........))))))...))))......)))))....))))))))))).... ( -31.30) >DroYak_CAF1 3598 93 - 1 UUCUCCUCGGAGGGGGGUGGCCCCAUUUAUUUUG------GGG-CUGAAACUGUUGGAGGGAAA--------AACAUAGGCGUAUAAAAAACCGGGUGGUGCCAAUGU (((.((((.((.((...((((((((.......))------)))-)))...)).)).))))))).--------.((((.(((.........(((....)))))).)))) ( -34.10) >DroAna_CAF1 4984 92 - 1 CGCUCUUCGG---GGGGUGGCCCCAUUUUUUAUUGUUCCAGCCGCCGAAAAUUUCAAU----AAAUAGA--AAACAAAA----CUAAAAAAUGGGGUGGUGCCAC--- ..(((....)---))(((((((((((((((((...(((........)))..((((...----.....))--))......----.)))))))))))))..))))..--- ( -24.50) >consensus UCCUCUUCGG___GGGGUGGCACCAUUUAUUUUG______GGGUCUGAAACUGUUGGAGGAAAAACAGC__AAACAGAAGCGUAUAAAAAACCGGGUGGUGCCAAUAU .((((........))))(((((((((((..(((..........((((...((((((.........))))))...))))..........)))..))))))))))).... (-12.15 = -16.40 + 4.25)


| Location | 9,045,787 – 9,045,880 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -21.29 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 9045787 93 + 22224390 CCAAAAAUAAAUGGUGCCACCCC---ACGAAGAGGAAAGAUUAGAAACCGGCGAAGGUUACGAUCGGGGGGGCCCGUUUCGCUUCUUUUUUUUUUA ........((((((.(((.((((---.(.....)....((((.(.((((......)))).))))))))).)))))))))................. ( -25.60) >DroSim_CAF1 3667 76 + 1 --CAAAAUAAAUGGUGCCACCCC---ACGAAGAGGAAAGAUUAGGAACCGGCGAAGGUUACGAUCAGGGGGGCCCGUUUCG--------------- --......((((((.(((.((((---.(.....)....((((...((((......))))..)))).)))))))))))))..--------------- ( -26.30) >DroYak_CAF1 3657 88 + 1 --CAAAAUAAAUGGGGCCACCCCCCUCCGAGGAGAAGAGAUUAGGAAACGGCGAAGGUCACGAUCAAGGGGGCCCGUUUCG------UUUUUUUCC --.((((((((((((....((((((((....)))....((((.(....)(((....)))..))))..)))))))))))).)------))))..... ( -30.40) >consensus __CAAAAUAAAUGGUGCCACCCC___ACGAAGAGGAAAGAUUAGGAACCGGCGAAGGUUACGAUCAGGGGGGCCCGUUUCG______UUUUUUU__ ........((((((.(((.((((....((..............(....)(((....))).))....)))))))))))))................. (-21.29 = -21.40 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:09 2006